Oncomedicine 2017; 2:71-79. doi:10.7150/oncm.18373 This volume Cite
Research Paper
Role of TRF2 in efficient DNA repair, spheroid formation and Cancer Stem Cell maintenance
1. Molecular Stress and Stem Cell Biology Group, School of Biotechnology, KIIT University, Bhubaneswar, Odisha-751024, India;
2. Department of Surgical Oncology, All India Institute of Medical Sciences, (AIIMS), Bhubaneswar, Odisha-751019, India;
3. Division of Research and Development, IMGENEX India Ltd, E-5, Infocity, Chandaka Industrial Estate, C S Pur, , Bhubaneshwar, Odisha- 751016.
*Contributed equally to this paper
Received 2016-11-16; Accepted 2017-1-13; Published 2017-3-10
Abstract
Chemotherapeutic resistance post-surgical management may be linked to an efficient DNA repair mechanism and survival of cancer stem cells. We report the presence of cancer stem cell like Side Population and their survival and enrichment post treatment with bleomycin in HCT116 cell line. The Side Population exhibited spheroidal characteristics and efficient repair of DNA damage foci compared to the parental HCT-116 cells. We report upregulated expression of β-catenin, TRF2, hTERT, CD44, CD24 and Oct4 genes in parental and clonospheres in a DNA damage environment. Comparative gene expression studies of parental and clonospheric HCT-116 cells revealed significant upregulation (p<0.01) of TRF2 gene in clonospheres. TRF2 which is a Telomere shelterin component is mainly involved in telomere maintenance and DNA repair. We report a role of TRF2 in cancer stem cell maintenance and efficient DNA repair in the clonospheric sub-population of HCT 116 cell line. Silencing of TRF2 gene significantly downregulated β-catenin, CD44, CD24, Oct4 and reduced the size of the clonospheres (4.448μm) when compared to scrambled non-silenced control (11.7273μm). TRF2 Silenced HCT116 parental cells reported compromised DNA repair and 2-fold increase in the number of DSBs as well as γH2Ax foci in the presence of a DSB generating agent Bleomycin, when compared to scrambled non-silenced control. In this study, we confirm a definitive role of TRF2 in stemness of cancer cells and DNA damage response and repair which is an extra-telomeric function and may have therapeutic implications in management of tumor recurrence.
Keywords: DNA Repair, TRF2, Clonospheres, Cancer Stem Cells, Stemness
Introduction
General clinical treatment regimen targets all cancer cells with an assumption that they have equal malignant potential. Current cancer treatment strategies are capable of targeting the bulk of the tumour cells but often fail to target the cancer stem cells, resulting in the recurrence of tumours [1].
Functional telomeres play a key role in maintenance of genome integrity, DNA damage response and repair [2, 3]. Functionality of the telomere in turn is maintained by a six member shelterin complex [4]. TRF2 (Telomerase Repeat binding Factor-2), an important shelterin component, not only protects the telomere [5, 6] but also exhibits extra-telomeric functions [7, 8]. TRF2 has been reported to play a major role in DSB repair [9, 10] where it interacts with repair proteins like MRN complex, Ku70, WRN and BLN [11]. Apart from involvement in DNA repair, TRF2, has been reported to interact with β-catenin which is a transcription factor and also a developmental gene [12]. However, not many reports are available regarding the mechanism of TRF2 mediated DNA damage response and its role in tumour stem cells.
Tumorspheres are three dimensional floating spheroid colonies grown in anchorage independent conditions in serum free medium [13, 14]. Tumorspheres, also known as tumour inducing cells, have been reported to exhibit properties of stem cells, like self- renewal, high proliferation rate, drug resistance properties and high tumorigenicity in vivo [13, 15, 16] along with expression of a higher percentage of stem cell markers like CD44 and ALDH1 [13, 14, 16]. Recently, tumorspheres derived from non-small cell lung cancer (NSCLC) cells showed altered DNA repair signalling possibly contributing to chemotherapy/radiotherapy resistance and subsequent maintenance of tumorigenicity [17].
There is another report by Bai et al which showed the role of TRF2 in maintenance of neurospheres in glioblastoma multiforme (GBM) [18].
In the current study we confirm a definitive role of TRF2 in maintaining stemness of cancer cells and efficient DNA damage response and repair in HCT-116 colon cancer cells in the presence of a Double Strand Break inducing drug, Bleomycin.
Materials and Methods
Reagents
Bleomycin was obtained from Biochem Pharmaceutical Industries Ltd and a working solution of 10mM in distilled water was prepared and stored at -20oC. HCT-116 (Homo sapiens colorectal carcinoma) was acquired from Imgenex Laboratories (Bhubaneshwar, Odisha, India) and maintained in DMEM (Himedia) supplemented with 10% (w/v) FBS (Himedia), and 100units/ml Penicillin-streptomycin-L Glutamine antibiotic (Himedia).
Clonospheres generation and maintenance
Single cell suspension of HCT-116 was plated in 6 well ultra-low attachment plates in serum deprived conditions in 37°C incubator with 5% CO2. Spheres formed after 10 days were collected by gentle centrifugation (Eppendorff) (700rpm, 5min), enzymatically dissociated with 0.5% Trypsin-EDTA (HiMEDIA) for 5min at 37°C, aspirated to form single cell suspension with a 22G needle (8 strokes) and sub-cultured at a density of 1200cells/ml in low attachment plates in DMEM (HiMEDIA) containing 1% antibiotic, supplemented with 0.1% FBS (HiMedia). For maintenance, media was changed once every eight days.
FACS analysis of Side Population and stem cell percentage
Bleomycin treated cells were harvested by trypsinization, washed twice and stained with Hoechst 33342 dye (final concentration 5μg/ml) and incubated at 37°C for 90 minutes. Cells were counter-stained with PI (1μg/ml) for 20 minutes at room temperature (RT). Suitable compensation controls for Hoechst and PI were taken. Confirmation of CD44 positive population post treatment was performed by incubating cell suspension with CD44 primary antibody (Abcam) (1:500 dilution) followed by FITC/TRITC tagged secondary antibody (Abcam) (1:1000 dilution). Flow cytometric analysis was performed using BDLSR Fortessa. Data was analysed using FlowJo software.
Table listing the primer sequences for β-catenin, hTERT, TRF2, CD44, CD24, Oct4 and β-actin genes used for real time quantitative PCR
| Genes | Forward Primer | Reverse Primer |
|---|---|---|
| β-catenin | 5'-GCTTTCAGTTGAGCTGACCA-3' | 5'- CAAGTCCAAGATCAGCAGTCTC-3' |
| hTERT | 5'-GCGTTTGGTGGATGATTTCT-3' | 5'- CAGGGCCTCGTCTTCTACAG-3' |
| TRF2 | 5'- GTACCCAAAGGCAAGTGGAA-3' | 5'- TGACCCACTCGCTTTCTTCT-3' |
| CD44 | 5'-CGGACACCATGGACAAGTTT-3' | 5'-CCGTCCGAGAGATGCTGTAG-3' |
| CD24 | 5'-GCTCCTACCCACGCAGATTT-3' | 5'-GAGACCACGAAGAGACTGGC-3' |
| Oct-4 | 5'-CCTGAAGCAGAAGAGGATCA-3' | 5'-CCGCAGCTTACACATGTTCT-3' |
| β-actin | 5'-TCACCCACACTGTGCCCATCTACGA-3' | 5'-CAGCGGAACCGCTCATTGCCAATGG-3' |
γ-H2Ax foci analysis
HCT-116 cells grown on coverslips were treated with Bleomycin (20-80µM) for 4 hrs followed by 2 hrs of recovery. Coverslips were washed, fixed in Methanol:Acetone (1:1), permeabilized by 0.25% PBST (1x PBS, 0.25% Triton-X100), blocked in 2.5% BSA in PBST for 1 hour at RT and probed overnight with γH2Ax primary antibody (1:2000 dilutions). FITC/TRITC fluorophore tagged secondary antibodies (1:4000 dilutions) was added for 1 hour at RT in dark. Coverslips were counterstained with 4, 6-diamidino-2-phenylindole (DAPI), mounted, sealed and observed. For clonospheres, single cell suspension post treatment were cytospun (Zettrek) on poly-l-lysine coated slides at 1100 rpm for 7 min at RT, fixed for 15 minutes with 4% paraformaldehyde and immunostained as mentioned above. Imaging was done using Olympus (BX 61) fluorescent microscopy and images were captured using ImagePro Express software. Number of γ-H2Ax foci/nuclei was counted using FociCounter software. A total of 100 nuclei was counted per concentration.
Single Cell Gel Electrophoresis (Comet Assay)
Treated cell suspensions were over-layered in agarose coated slides as described [19]. Gel electrophoresis was performed at 300mA current for 30 minutes at room temperature in dark. Slides were stained with PI (1μg/ml), cover slipped and imaging was done using Olympus (BX 61). Images were captured by ImagePro Express software. Comets were scored by CometScore software. Average olive tail moment [Olive Tail Moment = (Tail.mean - Head.mean) X Tail%DNA/100] was used to construct a comparative graph using GraphPad Prism 6 Software.
Real Time PCR
Real time PCR was performed by laboratory established protocol as described previously [20]. Primers used are mentioned in Table 1. Quantification was performed using the 2ΔΔCT method [21]. Statistical analysis was performed using the GraphPad Prism 6 software.
Selective gene silencing of shelterin component
Cells were transfected with DharmaFECT transcription reagent (Dharmacon) for transient siRNA silencing for a period of 48 hrs. Scrambled siRNA (siGENOME, Dharmacon) and TRF2 siRNA (siGENOME Human TERF2, Dharmacon) (Table 2) were used. Silencing was confirmed by RT-PCR analysis of extracted cDNA. Silenced cells were treated with Bleomycin and DSB signatures were analysed by γH2Ax assay as described earlier.
Table listing the siRNA used and the target sequences
| siRNA | Target Sequences |
|---|---|
| siGENOME Human TERF2 | GAAGUGGACUGUAGAAGAA |
| GGAAGCUGCUGUCAUUAUU | |
| GGAUCAGCUAUACAAUGUG | |
| GAAGACAGUACAACCAAUA | |
| SiGENOME Non-Targeting siRNA | UAGCGACUAAACACAUCAA |
Sphere formation assay
Transient TRF2 silencing was done in HCT-116 cells at 50-60% confluence as described previously. Scrambled siRNA was used as transfection control. Post silencing, cells were introduced to serum free environment and monitored for sphere formation. Images of Spheroids were captured by Olympus CKX41 inverted microscope. Spheroid size was calculated using automated particle size counting method by ImageJ software as described (http://mesa.ac.nz/2013/03/particle-sizing-using-imagej/). Number of spheres formed per 1000 cells seeded per well was also counted at 4 days and 7 days post seeding.
Immunocytochemistry
siTRF2 transfected cells were treated with increasing concentrations of Bleomycin sulphate (0µM, 40µM and 80µM) and immunocytochemistry was performed as per laboratory standardized protocols [22]. Cells were probed with CD44 primary antibody (1:2000 dilutions) overnight at 4°C followed by TRITC tagged secondary antibody (1:4000 dilutions) for 90 min at room temperature in dark. Slides were counterstained with DAPI (4, 6-diamidino-2-phenylindole), mounted, sealed and observed. Imaging was done by Olympus BX 61 Fluorescent microscope and images were captured by Image Pro Express software.
Statistical Analysis
Statistical analysis of One-way Analysis of Variance (ANOVA) and Two-way Analysis of Variance (ANOVA) was carried out for comparing two data groups using GraphPad Prism 6 software for all experiments conducted in duplicates. Unpaired student's t-test was used to compare average sphere sizes of silenced and control cells. Unpaired student's t-test was also used to compare gene expression variation in siTRF2 silenced versus non silenced scrambled siRNA HCT116 cells in the presence of bleomycin sulfate. All values of P<0.05 were considered as significant.
Results
Prevalence of a side-population of cells with stem cell like characters with increasing dosage of Bleomycin
A significant population of cells was observed to efflux Hoechst 33342 dye when treated with increasing concentration of Bleomycin (20, 40, 60, 80 μM) (Fig 1a and 1b). This small subpopulation of cells called Side Population (SP) increased with increasing concentration and then stabilized to a uniform percentage at higher concentrations. To further characterize the stem cell like properties of SP cells we treated HCT116 with increasing concentrations of Bleomycin and observed variation in CD44 percentage. Flow cytometry analysis revealed a significant increase in CD44 positive population (p<0.001) with increasing concentration of Bleomycin (20, 40, 60, 80 μM) when compared to untreated control (Fig 1c and 1d).
Increasing DNA damage as seen through γH2Ax immunocytochemistry (ICC)
To check for the extent of DSB generation, we treated HCT-116 with increasing concentrations of Bleomycin and quantified the formation of γH2Ax foci. Mean γH2Ax foci/nuclei was observed to significantly increase (p<0.05) with increasing concentrations of Bleomycin (20, 40, 60, 80 μM) in parental HCT-116 (Fig 1e and 1f). Increased foci are indicative of increasing DNA damage.
Comparative DNA damage profile and SP enrichment post Bleomycin sulphate treatment. a) Graph showing dose dependant (20, 40, 60, 80 µM) increase of number of enriched Side Population of HCT 116 post treatment with Bleomycin b) Representative FACS analysed FlowJo images showing enriched side population post treatment with Bleomycin. c) Graph showing dose dependant (20, 40, 60, 80 µM) increase in CD44 enriched cells post treatment with Bleomycin sulphate (p<0.001). d) Representative FACS image of enriched CD44 positive population post treatment with Bleomycin sulphate. e) Representative Immunocytochemistry (ICC) images of γH2Ax foci showing dose-dependent increase in DSB foci post treatment with Bleomycin treatment. f) Graph showing dose-dependent (20, 40, 60, 80 µM) increase in DSB foci post treatment with Bleomycin sulphate (p<0.05).
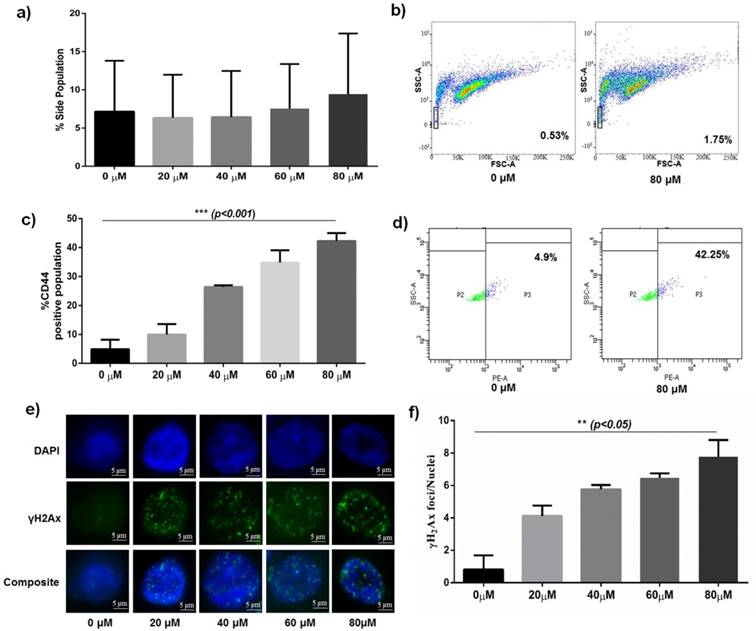
Differential gene expression in parental and clonospheric HCT116. a) Comparative images showing dose dependant γH2Ax foci formation in parental and clonospheric HCT116. b) Graph showing comparative frequency of γH2Ax foci/nuclei in parental and clonospheres (p<0.05). c) COMET showing DNA damage tails and Olive Tail Moment in 40 and 80 μM of dose in parental v/s clonospheric HCT 116. d) Graph showing quantitative average olive tail moment in parental and clonospheres post DNA damage with 40 and 80 μM of Bleomycin. e) Graph showing differential gene expression of TRF2, hTERT and β-catenin in parental HCT116 (p<0.001). f) Graph showing overexpression of TRF2, hTERT and β-catenin in clonospheric HCT116 (p<0.01).
Clonospheres treated with Bleomycin showed low γH2Ax signals than parental HCT116
To analyse the extent of DNA damage response (DDR) in resistant phenotype, we treated spheroidal clonospheres with specific concentrations of Bleomycin. With increasing dosage of Bleomycin, significantly low foci per nuclei (p<0.05) was observed in clonospheres as compared to that in parental HCT-116 (Fig 2a and 2b). Very low foci per nuclei indicate low DNA damage which is consistent with drug resistance.
Reduced COMET tail length in Bleomycin treated clonospheric HCT116
DNA damage, in terms of average olive tail moment, was observed to increase with increasing concentration of Bleomycin in Parental HCT 116 cells. No significant change in average olive tail moment was observed in clonospheres at the same concentrations (Fig 2c and 2d). Furthermore, tail length in clonospheres was significantly less than their parental counterpart (p<0.01) indicating less DNA damage.
Upregulated TRF2 expression in treated clonospheric HCT116
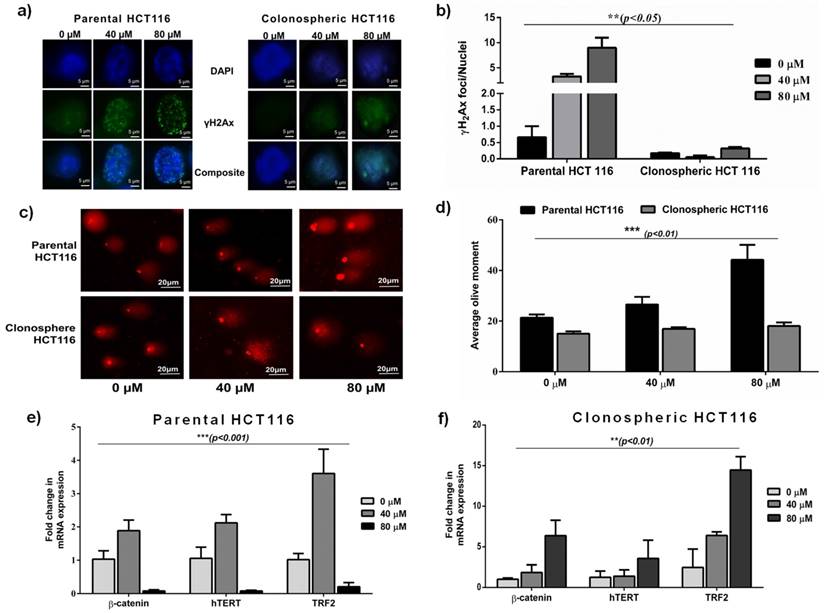
Comparative gene expression analysis revealed a significant overexpression of TRF2, β-catenin and hTERT genes (p<0.01) in clonospheres at high Bleomycin concentration (80μM) (Fig 2f). However in parental HCT116, TRF2 and related genes were observed to be significantly down regulated (p<0.001) (Fig 2e) at the same concentration (80μM). Corresponding to upregulated TRF2 expression at 80μM, stem cell markers like CD44 and Oct4 were also observed to be upregulated in Clonospheric HCT116 as compared to Parental HCT116 (Fig. 3a and 3b) Decreasing CD24 expression in clonospheric HCT116 was observed which correlated well with increased stem cell expression (Fig. 3b). Elevated TRF2 expression in clonospheres at high concentration of Bleomycin may contribute to increased expression of stem cell factors.
siTRF2 silenced HCT116 cells showed reduced repair capacity
Transient siTRF2 silencing was successfully achieved as shown in Fig 3c. Following which TRF2 silenced cells were treated with Bleomycin and γH2Ax assay was performed. With increasing concentration, a significant 2-fold increase in number of γH2Ax foci (green dots) in siTRF2 silenced HCT116 cells was observed compared to those formed in scrambled transfected HCT116 cells (p<0.01) (Fig 3d and 3e). Transient silencing of TRF2 hampers the DDR capacity of parental HCT116 cells.
siTRF2 silenced HCT116 cells also showed reduced stemness and sphere formation capacity
Differential gene expression showed significantly downregulated βcatenin, CD44, CD24 and Oct4 expression in siTRF2 silenced HCT116 cells (p<0.001) as compared to Scrambled transfected control (Fig. 3f). TRF2 silencing caused downregulation of developmental as well stem cell like factors.
Seven days post silencing, siTRF2 silenced cells showed significantly reduced average sphere size (~4.448μm) (Fig 3g) than Scrambled control (~11.7273μm) (p<0.0001). Number of spheres formed per 1000 cells seeded was also significantly lower in siTRF2 silenced cells than in the scrambled (p<0.05) (Fig 3h). Phase contrast microscopic images of spheres at Day 4 and Day 7 post silencing shows larger size and higher number of spheres in scrambled than in siTRF2 silenced HCT116 (Fig 3i).
DNA damage profile and reduced sphere formation in siTRF2 silenced HCT116. a) Graph showing differential stem cell expression (CD44, CD24 and Oct4) in Bleomycin treated Parental HCT116 (p<0.001) b) Graph showing significant overexpression (p<0.0001) of CD44, Oct4 and downregulation of CD24 in clonospheric HCT116 cells with increasing concentration of Bleomycin (0µM, 40µM, 80µM). c) Graph showing expression of TRF2 post silencing in siTRF2 transfected cells respect to Scrambled siRNA transfected cells in HCT 116 (p<0.05). d) Graph showing higher frequency of γH2Ax foci/nuclei at 40 and 80 μM dose of Bleomycin post silencing in siTRF2 silenced cells compared to Scrambled transfected HCT-116 cells (p<0.05). e) Representative images of γH2Ax foci/nuclei at 40 and 80 μM dose of Bleomycin post silencing in siTRF2 silenced cells compared to Scrambled HCT116. f) Differential gene expression showing significant downregulation of βcatenin, CD44, CD24 and Oct4 genes in siTRF2 silenced HCT116 cells as compared to scrambled transfected HCT116 cells (p<0.005). g) Graph showing significant variation (p<0.0001) in the average size of the spheres in siTRF2 transfected cells (~4.4µm) when compared to scrambled (~11.72μm). h) Graph showing reduced number of spheres formed in siTRF2 silenced cells after 7 days post silencing as compared to Scrambled transfected HCT116 (p<0.05). i) Representative phase contrast microscopic images at 40x magnification showing comparative size of spheres formed in siTRF2 silenced cells as compared to Scrambled.
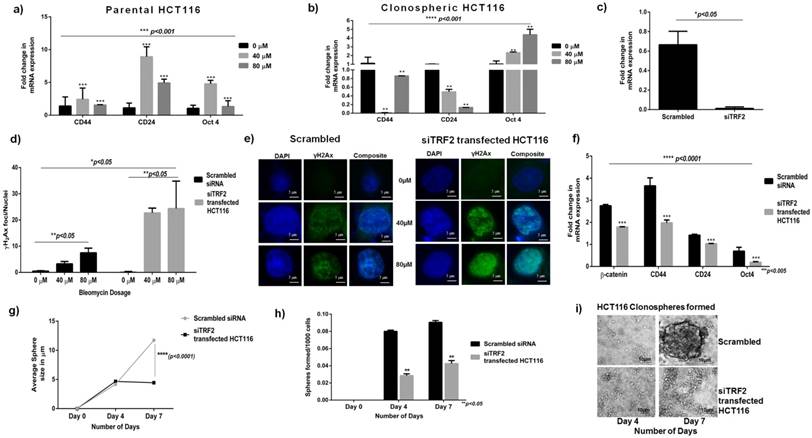
Reduced stem cell like population in absence of TRF2 expression when exposed to Bleomycin: a) Graph showing significant downregulation of β-catenin in Bleomycin treated siTRF2 silenced HCT116 cells as compared to Bleomycin treated scrambled transfected cells (p<0.05). b) Graph showing significant downregulation of CD44 in Bleomycin treated siTRF2 silenced HCT116 cells as compared to Bleomycin treated scrambled transfected cells (p<0.05). c) Graph showing significant downregulation of CD24 in Bleomycin treated siTRF2 silenced HCT116 cells as compared to Bleomycin treated scrambled transfected cells at 80µM concentration (p<0.05). d) Immunocytochemistry analysis showing reduced CD44 expression in siTRF2 silenced HCT116 post treatment with Bleomycin sulphate when compared to scrambled transfected HCT116 post treatment with Bleomycin.
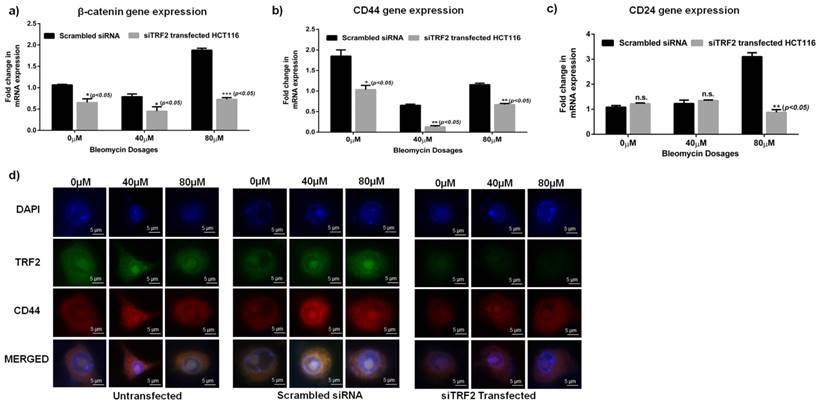
Furthermore, siTRF2 silenced HCT116 cells when treated with increasing concentrations of Bleomycin (0μM, 40μM, 80μM) showed a significant downregulation of CD44 (p<0.05) and β-catenin (p<0.05) genes as compared to Scrambled transfected HCT116 cells (Fig 4 a-c). CD24 was also significantly reduced in siTRF2 transfected cells at 80µM of Bleomycin dosage (Fig. 4c). Immunocytochemistry also confirmed reduced expression of CD44 in siTRF2 silenced HCT116 cells as compared to Scrambled siRNA when treated with increasing concentration of Bleomycin (Fig 4d).
Discussion
Solid tumours are reported to possess a sub set of cells called Side Population expressing various stem like cell markers, self-renewal capacity and ability to give rise to differentiated cells [23, 24]. These resistant populations undergo various cellular and genetic adaptations such as dormancy, efficient DNA repair mechanism, overexpression of multi drug resistance transporters, surpassing apoptosis and drug inactivation or its efflux from the cell [25]. To check for the presence of this subpopulation of SP cells, we exposed HCT116 to increasing concentrations of Bleomycin and performed Hoechst efflux assay. Bleomycin is a DSB inducing drug that has been widely used as a chemotherapeutic agent [26]. An increase in the percentage of Hoechst negative cells were observed with increasing concentration of Bleomycin (Fig 1a and 1b). Presence of a biologically significant amount of SP cells in the parental population is indicative of the presence of cells with efficient DNA repair capacity. A significant increase in stem cells in terms of CD44 positive population was observed with higher Bleomycin concentration (p<0.001) (Fig 1c and 1d) . This indicates exposure to DNA damaging environment induces enrichment of stem cell like population. Measuring the number of γH2Ax foci per nucleus gives the extent of DSB sites formed on exposure to DSB generating agent [27]. As shown in Fig 1f, significant increase in γH2Ax foci per nucleus in higher concentration of bleomycin treated HCT116 indicates the presence of multiple DSB sites in parental HCT-116 (p<0.05).
Following the DSB generation in parental, we compared the DSB generation profile in spheroids (clonospheres). Response of clonospheres was observed only at 40 and 80µM concentration of bleomycin sulphate. Hence, for further analysis, the drug concentrations of 40 and 80µM were used. On treatment with specific concentrations (40 and 80µM) of the drug, the frequency of γH2Ax foci formed in clonospheres was also significantly lower than those formed in parental HCT-116 (p<0.05) as shown in Fig 2b. Low number of γH2Ax foci formed/clonospheric nuclei post exposure to high concentrations (80μM) of bleomycin is indicative of rapid repair of DSBs. The average olive tail moment obtained by COMET assay was significantly less in clonospheres when compared to their parental counterpart (p<0.001) (Fig 2d). To our knowledge, this is the first direct report of efficient DSB repair in cancer stem cell clonospheres.
β-catenin, a transcription factor, has been known to crosstalk with several developmental pathways [28] and is reported to play a role in DNA damage repair [29, 30]. Previously we reported importance of β-catenin in prognosis of oral cancer and its overexpression in clonospheres along with hTERT and TRF2 [20, 22]. In the present study, we observed a significant overexpression of β-catenin, TRF2 and hTERT at 40µM concentration and low expression was observed at 80µM concentrations compared to the untreated control (p<0.001) in parental HCT-116 (Fig 2e). In clonospheres, an overexpression of all the genes was observed at increasing concentration of Bleomycin compared to the untreated control (p<0.01) as seen in Fig 2f. Amongst the genes considered, TRF2 was seen to correlate well with the DNA damage status of parental and clonospheric HCT-116. Low TRF2 expression was observed at a concentration of 80μM (Fig 2e) whereas high DSB was observed in the same concentration in parental HCT-116 (Fig 2b). In contrast, high TRF2 expression observed at higher concentration (80μM) in clonospheres (Fig 2f) corresponded to the presence of low DSB foci in clonospheres (Fig 2b). Since TRF2 is involved in DSB repair [9, 10], TRF2 overexpression in clonospheres is indicative of its involvement in an efficient damage repair mechanism. Furthermore, on treatment with 80μM bleomycin, a significant (p<0.05) increase in expression of stemness genes like CD44, Oct 4 and a significant downregulation of CD24 gene (p<0.05) was observed in clonospheric HCT116 when compared to Parental HCT116 (Fig. 3a and 3b). CD44, CD24 are both stem cell markers that are linked to spheroidal phenotype [31-33]. High CD44 coupled with low CD24 expression is indicative of stem cell phenotype in clonospheres. Oct-4 is a transcription factor which has been reported to be involved in various cancers and is also marker for cancer stem cell like population [34, 35]. High expression of TRF2 also corresponded with increased stem cell like expression of CD44 and Oct 4 in clonospheres which is indicative of the involvement of TRF2 in CSC maintenance.
To confirm the importance of TRF2 in imparting efficient DSB repair and CSC maintenance, we silenced TRF2 in parental HCT-116 (Fig 3c) and observed DSB repair response. γH2Ax assay revealed a significant increase (p<0.05) in number of foci/nuclei in siTRF2 cells compared to scrambled (Fig 3d and 3e). Absence of TRF2 resulted in increased DSB sites which led us to conclude that TRF2 is indispensable for efficient DSB repair in cancer cells. A significant (p<0.005) downregulation of CD44 and Oct4 was also observed in siTRF2 treated cells compared to untreated Scrambled HCT116 (Fig 3f). Apart from the stem cell markers, β-catenin was also observed to be downregulated in siTRF2 silenced cells (Fig. 3f). Furthermore, when subjected to sphere formation, siTRF2 transfected HCT 116 cells formed significantly smaller (p<0.0001) and fewer number of spheres compared to that of scrambled (Fig 3g-i) over a period of 7 days. β-catenin, as a transcription factor is reported to interact with TRF2 [12] and is also reported to be involved in maintenance of spheroidal phenotype [14, 36]. Loss of function of TRF2 resulted in decreased β-catenin expression which was reflected in reduced sphere formation efficiency and decreased spheroid size over a period of 7 days. With the absence of TRF2, stem cell like characters in terms of CD44, CD24 and Oct4 were also downregulated which correlated well with reduced spheroidal population. To confirm this observation, on treatment of siTRF2 HCT116 cells with increasing Bleomycin concentrations (0μM, 40 μM and 80 μM), we observed a significantly reduced expression of CD44 and β-catenin genes compared to that of scrambled transfected HCT116 cells (Fig. 4a-c). At 80µM concentration, significantly reduced CD24 expression was observed (p<0.05) in siTRF2 transfected HCT116 as compared to Scrambled siRNA transfected cells. Silencing of TRF2 results in downregulation of CD24 along with CD44 and β-catenin in the presence of a DNA damaging environment. Reduced expression of CD44 was also observed by immunofluorescence in Bleomycin treated siTRF2 transfected cells (Fig. 4D). Recently, TRF2 deficiency was reported to radiosensitize human Mesenchymal Stem cells through destabilization of the telomere [37]. In the present work, our observations led us to conclude that inhibition of TRF2 abrogates the formation and maintenance of the spheroidal phenotype and loss of TRF2 results in down regulation of CSC characteristics when exposed to DNA damaging environment in HCT116 colon cancer cell line. Hence TRF2 expression is indispensable for spheroid formation and CSC maintenance.
In conclusion, we speculate that functional TRF2 is involved in an extra-telomeric activity of imparting efficient DSB repair in the spheroidal population in the presence of a DNA damaging environment. Furthermore, expression of TRF2 also plays an integral part in the formation and maintenance of the spheroidal phenotype including enrichment of the CSC phenotype, the mechanism of which needs to be elucidated.
Acknowledgements
This work was supported by extramural funding from Department of Biotechnology (DBT), Government of India (Grant no.:BT/PR/14659/MED/31/108/2010) and Board of Research in Nuclear Science, Department of Atomic Energy, Government of India (Grant No.:2013/35/45/BRNS). We also acknowledge the technical support of the MSSB (Molecular Stress and Stem Cell Biology) group, School of Biotechnology.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lobo NA, Shimono Y, Qian D, Clarke MF. The Biology of Cancer Stem Cells. Annual Review of Cell and Developmental Biology. 2007;23:675-99
2. Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nature reviews Genetics. 2005;6:611-22
3. Doksani Y, de Lange T. The role of double-strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harbor perspectives in biology. 2014;6:a016576
4. Lange Td. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19(18):2100-10
5. Feuerhahn S, Chen L-y, Luke B, Porro A. No DDRama at chromosome ends: TRF2 takes centre stage. Trends in Biochemical Sciences. 2015;40:275-85
6. Rai R, Chen Y, Lei M, Chang S. TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nature Communications. 2016;7:10881
7. Blasco PM, María A. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nature Reviews Cancer. 2011;11:161-76
8. Mao Z, Seluanov A, Jiang Y, Gorbunova V. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13068-73
9. Huda N, Abe S, Gu L, Mendonca MS, Mohanty S, Gilley D. Recruitment of TRF2 to laser-induced DNA damage sites. Free Radical Biology and Medicine. 2012;53:1192-7
10. Huda N, Tanaka H, Mendonca MS, Gilley D. DNA damage-induced phosphorylation of TRF2 is required for the fast pathway of DNA double-strand break repair. Molecular and cellular biology. 2009;29:3597-604
11. Bradshaw PS, Stavropoulos DJ, Meyn MS. Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nature genetics. 2005;37:193-7
12. Diala I, Wagner N, Magdinier F, Shkreli M, Sirakov M, Bauwens S. et al. Telomere protection and TRF2 expression are enhanced by the canonical Wnt signalling pathway. EMBO Rep. 2013;14:356-63
13. Sukach AN, Ivanov EN. Formation of spherical colonies as a property of stem cells. Cell and Tissue Biology. 2007;1:476-81
14. Kanwar SS, Yu Y, Nautiyal J, Patel BB, Majumdar APN. The Wnt/beta-catenin pathway regulates growth and maintenance of colonospheres. Molecular cancer. 2010;9:212
15. Yeung TM, Gandhi SC, Wilding JL, Muschel R, Bodmer WF. Cancer stem cells from colorectal cancer-derived cell lines. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3722-7
16. Qiu X, Wang Z, Li Y, Miao Y, Ren Y, Luan Y. Characterization of sphere-forming cells with stem-like properties from the small cell lung cancer cell line H446. Cancer letters. 2012;323:161-70
17. Lundholm L, Hååg P, Zong D, Juntti T, Mörk B, Lewensohn R. et al. Resistance to DNA-damaging treatment in non-small cell lung cancer tumor-initiating cells involves reduced DNA-PK/ATM activation and diminished cell cycle arrest. Cell death & disease. 2013;4:e478
18. Bai Y, Lathia JD, Zhang P, Flavahan W, Rich JN, Mattson MP. Molecular Targeting of TRF2 Suppresses the Growth and Tumorigenesis of Glioblastoma Stem Cells. Glia. 2014;62:1687-98
19. Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Experimental cell research. 1988;175:184-91
20. Padhi S, Saha A, Kar M, Ghosh C, Adhya A, Baisakh M. et al. Clinico-Pathological Correlation of β-Catenin and Telomere Dysfunction in Head and Neck Squamous Cell Carcinoma Patients. J Cancer. 2015;6:192-202
21. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402-8
22. Saha A, Shree Padhi S, Roy S, Banerjee B. HCT116 colonospheres shows elevated expression of hTERT and β-catenin protein - a short report. Journal of stem cells. 2014;9:243-51
23. Yao H, Ashihara E, Maekawa T. Targeting the Wnt/β-catenin signaling pathway in human cancers. Expert opinion on therapeutic targets. 2011;15:873-87
24. Song J, Chang I, Chen Z, Kang M, Wang C-Y. Characterization of side populations in HNSCC: highly invasive, chemoresistant and abnormal Wnt signaling. PloS one. 2010;5:e11456
25. Hu Y, Fu L. Targeting cancer stem cells: a new therapy to cure cancer patients. American journal of cancer research. 2012;2:340-56
26. Chen J, Ghorai MK, Kenney G, Stubbe J. Mechanistic studies on bleomycin-mediated DNA damage: multiple binding modes can result in double-stranded DNA cleavage. Nucleic acids research. 2008;36:3781-90
27. Sánchez-Flores M, Pásaro E, Bonassi S, Laffon B, Valdiglesias V. γH2AX assay as DNA damage biomarker for human population studies: defining experimental conditions. Toxicological sciences: an official journal of the Society of Toxicology. 2015;144:406-13
28. Bertrand FE, Angus CW, Partis WJ, Sigounas G. Developmental pathways in colon cancer: Crosstalk between WNT, BMP, Hedgehog and Notch. Cell Cycle. 2012;11:4344-51
29. Chang HW, Nam HY, Kim HJ, Moon SY, Kim MR, Lee M. et al. Effect of beta-catenin silencing in overcoming radioresistance of head and neck cancer cells by antagonizing the effects of AMPK on Ku70/Ku80. Head & neck. 2016;38(Suppl 1):E1909-17
30. Dose M, Emmanuel AO, Chaumeil J, Zhang J, Sun T, Germar K. et al. β-Catenin induces T-cell transformation by promoting genomic instability. Proc Natl Acad Sci U S A. 2014;111:391-6
31. Cao L, Zhou Y, Zhai B, Liao J, Xu W, Zhang R. et al. Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC gastroenterology. 2011;11:71
32. Elkord AJaE. Significance of CD44 and CD24 as Cancer Stem Cell Markers: An Enduring Ambiguity. Clinical and Developmental Immunology. 2012;2012:11
33. Sahlberg SH, Spiegelberg D, Glimelius B, Stenerlöw B, Nestor M. Evaluation of Cancer Stem Cell Markers CD133, CD44, CD24: Association with AKT Isoforms and Radiation Resistance in Colon Cancer Cells. PLoS ONE. 2014;9:e94621
34. Tsai L-L, Hu F-W, Lee S-S, Yu C-H, Yu C-C, Chang Y-C. Oct4 Mediates Tumor Initiating Properties in Oral Squamous Cell Carcinomas through the Regulation of Epithelial-Mesenchymal Transition. PLoS ONE. 2014;9:e87207
35. Liu J, Wang LEI, Ma L, Xu J, Liu C, Zhang J. et al. Significantly increased expression of OCT4 and ABCG2 in spheroid body-forming cells of the human gastric cancer MKN-45 cell line. Oncology Letters. 2013;6:891-6
36. Cai C, Zhu X. The Wnt/beta-catenin pathway regulates self-renewal of cancer stem-like cells in human gastric cancer. Mol Med Rep. 2012;5:1191-6
37. Orun O, Tiber PM, Serakinci N. Partial knockdown of TRF2 increase radiosensitivity of human mesenchymal stem cells. International journal of biological macromolecules. 2016;90:53-8
Author contact
![]() Corresponding author: Birendranath Banerjee, Group leader, Molecular stress and stem cell biology group, Disease Biology Lab, KIIT School of Biotechnology, KIIT University, Bhubaneshwar-751024, Odisha, India: Email: bnbanerjeeac.in, phone: +91-9090840042 Fax: 0674-2378776
Corresponding author: Birendranath Banerjee, Group leader, Molecular stress and stem cell biology group, Disease Biology Lab, KIIT School of Biotechnology, KIIT University, Bhubaneshwar-751024, Odisha, India: Email: bnbanerjeeac.in, phone: +91-9090840042 Fax: 0674-2378776