Oncomedicine 2018; 3:82-93. doi:10.7150/oncm.25573 This volume Cite
Review
Impact of Host Molecular Genetic Variations and HIV/HPV Co-infection on Cervical Cancer Progression: A Systematic review
1. MRC Unit for Genomic and Precision Medicine, Division of Human Genetics, Department of Pathology, Institute of Infectious Disease and Molecular Medicine Faculty of Health Sciences, University of Cape Town, South Africa
2. Department of Gynaecology, Morogoro Regional Referral Hospital, Morogoro, Tanzania
3. Division of Immunology, Department of Pathology, Institute of Infectious Disease and Molecular Medicine, Faculty of Health Sciences, University of Cape Town, South Africa
4. Laboratory for Tissue Immunology, National Health Laboratory Services, Groote Schuur Hospital, Cape Town, South Africa
5. Division of Medical Virology, Department of Pathology, Faculty of Health Sciences, University of Cape Town
6. Department of Biochemistry and Medical Microbiology, University of Namibia School of Medicine, Windhoek, Namibia.
Received 2018-2-15; Accepted 2018-7-29; Published 2018-9-4
Abstract
Only a small subset of women who are co-infected with Human Immunodeficiency Virus sub-type 1 (HIV) and persistence oncogenic Human papillomavirus (HPV), progress rapidly to invasive cervical cancer by mechanisms that are currently poorly understood. The use of Highly Active Antiretroviral Therapy (HAART), with ensuing immune reconstitution of CD4 T-cells, does not appear to prevent rapidly progressing cervical carcinogenesis. Therefore, to better understand the cervical cancer pathogenesis in HIV/HPV co-infected women, this review focuses on identifying host molecular genetic variations and genetic alterations in cervical cancer progression that may play a role in disease progression. This is an important aspect for individualised genomic profiling and targeted molecular prevention in order to improve the management of the disease in this sub-population.
Keywords: Host molecular genetics, cervical cancer, HIV/HPV co-infection, genomic profiling and molecular targeted prevention
1. Introduction
Every year, more than 530,000 women worldwide are diagnosed with invasive cancer of the uterine cervix and approximately 275,000 die from the disease (1, 2). More than 88% of these deaths occur in developing countries especially in sub Saharan Africa (3).
Oncogenic Human papillomavirus (HPV), is one of the primary causative agents for cervical cancer (4). However, Human Immunodeficiency Virus sub-type 1 (HIV) as a co-infection, may influence cervical disease progression and invasive cancer outcomes in HIV positive women (3, 5-8). HIV positive women are also more likely to have concurrent infections of single or multiple strains of oncogenic HPV, compared to HIV negative women (4, 6, 9-12). Furthermore, published data clearly shows that, persistent oncogenic HPV and HIV co-infections, contribute to rapidly progressing cervical carcinogenesis when compared to HIV negative women with a single, multiple, or no HPV infection (13-16). Although HIV/HPV co-infection is common in sub-Saharan Africa, only relatively few infected patients develop cervical disease and only about one third of 'in situ' cervical carcinomas progress to invasive cancer (9, 11). Surprisingly, the risk of developing invasive cervical cancer does not decrease following Highly Active Antiretroviral Therapy (HAART), which targets the HIV infection (9, 17-20). On the contrary, a proportion of the patients with invasive cervical cancer have no detectable HIV/HPV viral co-infection (21, 22).
It has been argued that, despite underlying immunodeficiency and immune reconstitution, the existing cervical carcinogenesis process in HIV/HPV co-infected women is further influenced by host molecular genetic factors, which vary between individuals (18, 23, 24) (Figure 1). It has also been suggested that the rate of cervical disease progression and likely protection may depend on host immunogenetic variations (25-29).
Cervical carcinogenesis process in HIV/HPV co-infected women. SIL is squamous intraepithelial lesion.
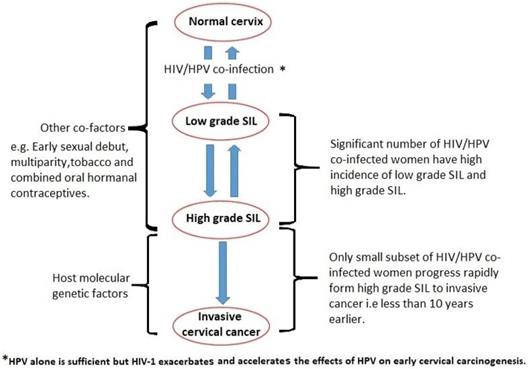
Specifically, the argument is that, cervical cancer progression is controlled via the microsatellite instability pathway amongst HIV/HPV co-infected women and through the loss of heterozygosity (LOH) pathway amongst HPV-only infected women (18, 23, 30, 31). While many studies have documented the association between HIV infection and rapidly invasive cervical cancer development, none have shown a direct link between HIV/HPV co-infection and the degree of disease invasiveness and rate of progression (32-34).
These considerations have determined the focus of this review which is to assess the current knowledge of the influence of host molecular genetic variations, genetic alterations, and HIV/HPV co-infection on rapidly progressing cervical carcinogenesis. We have reviewed published cohort studies and further investigations. We have also examined the evidence for possible involvement of host molecular genetic alterations and immunogenetic variations in HIV/HPV co-infected cervical carcinogenesis. This review stems from research conducted into further knowledge on individualised genomic profiling and targeted molecular prevention in order to improve management of the disease in this subpopulation (8, 35).
2. Methods
We have conducted our methodology according to similar published studies (36, 37). A comprehensive, systematic literature search of peer-reviewed, published articles from the NCBI, PubMed, EBSCO, Medline, Elsevier Science, Springer Link and the Google Scholar bibliographic databases was carried out. We included all original research studies, short communications, critical reviews and meta-analyses, reports on genes susceptible for cervical cancer and immune response, and factors changing risk or offering likely protection of cervical cancer development, ever published prior to May, 31st 2018. Information on specific alleles and/or genes were noted and critically reviewed. The key words/phrases used for the search were 'host molecular genetics', 'cervical cancer susceptibility', 'HIV/HPV co-infection', 'HIV and cervical cancer', 'cancer genomic profiling and molecular targeted prevention', 'cervical cancer, mutations and viruses' and 'cervical cancer genetics'. We excluded all studies on epigenetics of cervical cancer as these were not deemed to be relevant to our study topic and focus. There were no specific analytical methods used for the examination of articles due to the nature of our study.
3. Results and Discussion
In this section we will discuss;
i) Cervical cancer pathogenesis, HAART and HIV/HPV co-infection.
ii) Host HLA polymorphisms, genetic mutations and HIV/HPV co-infection in cervical cancer progression.
iii) Molecular genetic variations, genes, chromosomes, SNPs and HPV persistence in cervical disease progression and susceptibility.
iv) What is not known in host molecular genetics of cervical carcinoma?
i) Cervical cancer pathogenesis, HAART and HIV/HPV co-infection
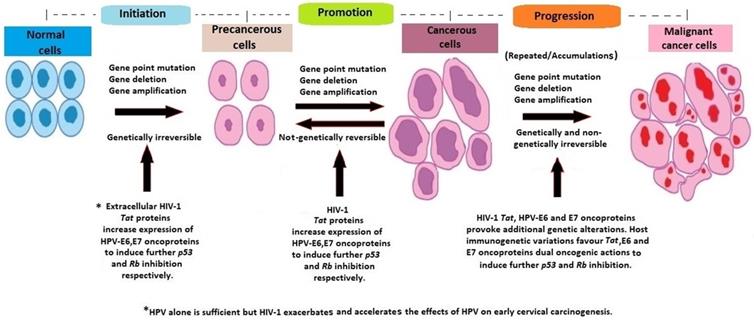
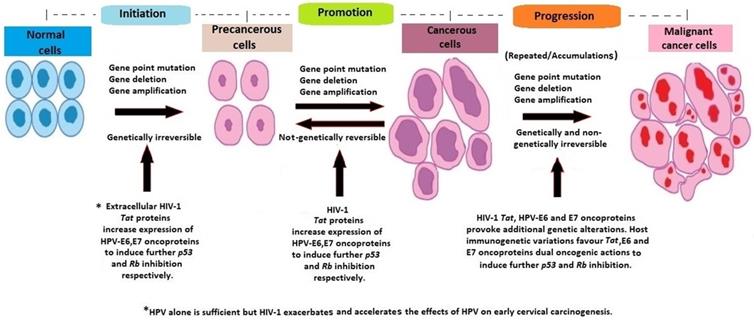
The pathogenesis of cervical cancer involves three important steps; high risk HPV viral genome integration into host genome, the oncogenic effects of HPV oncoproteins E6, E7, and the accumulation of recurrent, unrepaired genetic alterations in host chromosomal DNA (38-41). Furthermore, there is an interaction between HPV and HIV co-infections, resulting in an increased risk of HPV-associated morbidity and cervical cancer mortality among HIV-positive women (34, 42). In a few cases, HIV/HPV-associated cervical cancerous cells, non-genetically regress spontaneously to pre-cancerous cells. Persistent accumulation of uncorrected mutations and additional pro-oncogenic effects of HIV-1 Tat and HPV E6, E7 oncoproteins, lead to the progression of cancerous cells to invasive malignant cancer cells, regardless of the use of HAART (11, 14, 34, 43, 44) (Figure 2).
Host molecular genetic variations and alterations in HIV/HPV co-infected cervical carcinogenesis.
The immunosuppressive effects of HIV infection are associated with the rapid progression of HPV-induced cervical pre-malignant lesions. This may influence the rapid onset of cervical disease and further effects on clinical outcome. However, the direct mechanism is not clear (45-47). Additionally, the effects of duration of HAART use in cervical carcinogenesis prevention are still unknown (48, 49).
In a study by De Jong et al (50), they suggested that absence of functional HPV16-specific CD4+ T-cell immune responses found in women with cervical cancer may explain further development of the cervical disease despite immune reconstitution following HAART initiation or in patients with competent CD4+ T-cell count. Additionally, it has been already reported that not all women who progress rapidly to invasive cervical cancer are HIV positive. This suggests that, there are many other possible host factors apart from immunosuppression or HIV/HPV co-infection that may play a role in rapidly progressing cervical carcinogenesis (39, 42, 51, 52).
HIV proteins can directly cause cancer growth by interfering with cellular functions (8, 42, 49, 53). For example, HIV Tat proteins directly interact with the host tumour suppressor genes p53/pRb/p130/p107 and induce increased cell proliferation, which promote the effect of HPV oncoproteins E6 and E7 in the rapidly progressing cervical carcinogenesis (33, 42, 49, 53, 54). The increased rate of HPV-associated cervical disease in HIV positive women is aggravated by HIV/HPV molecular interactions because HIV Tat proteins can modulate HPV E2 gene expression, which in turn, influences HPV viral replication (55, 56). However, Hirbod et al. (57), Nkwanyana et al. (58) and Bebell et al. (59) have suggested that cervical mucosal inflammation in HIV infected women may be associated with low CD4 (+) cell counts during acute infection. Although this suggestion agrees with cancer microenvironment theories that chronic and persistent inflammation contributes to cancer development and can predispose to rapidly progressing cervical carcinogenesis, the effect has been observed only in a subset of HIV/HPV co-infected women (60-62). It has been shown that, inflammatory mediators on the surface of the uterine cervical epithelial cells, can promote adhesion of HIV infected leukocytes. This in turn facilitates HIV Tat proteins uptake by cervical epithelial cells before their transformation into cancerous cells (54).
The oncogenic potential of high risk HPV may contribute to the accumulation of mutations in proto-oncogenes and tumor suppressor genes between G1 phase and S-phase of the cell cycle in the host (23, 63). This process is mostly mediated by the oncoproteins E6 and E7 which hijack host genomic DNA, then bind and inactivate the tumour suppressor genes, p53 and the retinoblastoma protein (pRb), respectively, to further inhibit apoptosis. This process is achieved through the Ubiquitin-mediated degradation pathway (13, 64-66).
It is clear therefore that, apart from immunosuppression, additional genetic and genomic alterations are necessary for pre-cancerous cells to sufficiently progress into malignant and invasive cancer cells, especially following HAART, which reconstitute the host immune competency (67).
ii) Host HLA polymorphisms, genetic mutations and HIV/HPV co-infection in cervical cancer disease progression
In some populations, carcinogenesis of squamous cell carcinoma is more susceptible to genetic variations and alterations than adenocarcinoma (68, 69). Since the immune system normally mobilizes to clear viral infections, the development of virus-associated cancers result from a failure of anti-viral immunity. Furthermore, in cervical carcinogenesis, host immune-competence gradually diminishes as cervical disease progresses to invasive cervical cancer (64).
The genes coding for the Human Leukocyte Antigen (HLA) system, which reside on the short arm of human chromosome 6, (except for the gene for β2-microglobulin) play a central role in immune recognition and the subsequent clearance of virally infected cells (70-75). Therefore, any variations or structural genomic changes within the HLA region, which influence immune evasion by viruses and cancer cells, may determine the lesions likely to progress to invasive cancer (43, 49, 71, 73, 76). In a study of HLA genes in cervical cancer patients compared to healthy controls, expression of HLA I genes, were downregulated, suggesting that host genetic variations of HLA class I genes also have a significant bearing on specifically HPV16-related cervical carcinogenesis (77). HPV16 E5 oncoprotein and downregulation of the surface HLA class I expression has already been reported (78). However, genetic predisposition and variability in the HLA genes, have shown considerable contradictory findings in different study populations (36, 79, 80). Variations in HLA II genes and cervical cancer susceptibility have been investigated in different geographical populations with inconsistent findings, highlighting the complexity of the viral/host/environmental ecosystem (69, 80-85). For example, predispositions of the HLA system and a positive cervical cancer association with the TNF genes for apoptosis was previously reported (86-88). Although in the South African population, TNF G-308 did not show any association with cervical cancer susceptibility (37, 89).
It has been widely reported that loss of heterozygosity (LOH) mutations and microsatellite instability (MSI) at multiple genomic loci including the HLA region, are the most common host genetic alterations seen in cervical cancer tissues (90, 91). This possibly indicates that the integrity of the HLA region is compromised during the process of carcinogenesis (92-94). LOH and MSI are caused by genetic alterations such as the physical deletion of a chromosomal region, a tumor suppressor gene, or chromosomal non-disjunction during mitotic recombination. It has been reported that the mechanisms of LOH/MSI may be remarkably chromosome-specific (73, 90, 93, 95, 96), with some chromosomes being completely lost while more than half of the losses are associated with the loss of only a part of the chromosome (90, 97). Essentially, the mechanism underlying HIV/HPV co-infection and cervical cancer development with regard to LOH/MSI or immunogenetic variations is still poorly understood (72). It has been hypothesised that, cervical cancer progression is influenced by the extent of LOH/MSI and HLA variations in immune response towards oncogenic HPV clearance and the combined pro-oncogenic effects of the HIV/HPV co-infection (96, 98-100).
Host genetic variations in the HLA genes that influence the primary immune response and the severity of LOH/MSI at the HLA genomic loci may, therefore, determine which lesions are at the highest risk for rapid progression to invasive cervical cancer in HIV/HPV co-infected women (36, 79).
iii) Molecular genetic variations, genes, chromosomes, SNPs and HPV persistence in cervical disease progression and susceptibility
Currently, there are no consistent data on the association between any gene polymorphism in cervical cancer and disease outcome (101). In studies of different chromosomes, the changes involving loss of 2q, 3p, 4p, 4q, 5q, 6q, 11q, 13q, 17q and 18q regions and gain of 1q, 3q, 5p and 8q at various stages of cervical cancer have shown possible association with either oncogenic HPV persistence or cervical cancer disease progression (63, 102) (Table 1). Presence of chromosomal aneuploidy, which increases genomic instability in rapidly progressing carcinogenesis was reported in both cervical pre-cancer and cervical invasive cancer (103-105). Furthermore, the presence of an isochromosome 5p associated with cervical cancer susceptibility has been reported in a number of different studies (106-108).
Summary of host gene polymorphisms, SNPs and chromosomal locations found to be associated with either HPV persistence or cervical disease progression worldwide.
| HOST GENES AND SNPs | CHROMOSOME | COHORT SIZE AND NATURE OF THE STUDY | POPULATION STUDIED | HPV 16 OR 18 GENOME INTERACTION WITH HOST GENES |
|---|---|---|---|---|
| A) Genes involved in HPV persistence | ||||
| CTLA-4,-318 C/T | 2q33.2 | Case-control, 144 cases vs 378 controls | Taiwan (147) | - |
| STING | 5q31.2 | Cross-sectional, 148 patients | Thailand | E2 downregulates STING (148) |
| HLA-DQB1 | 6p21.32 | Case-control, 1306 cases vs 288 controls | Sweden (82) | E5, E7 downregulate expression of MHC molecules (149) |
| GTF2H4 rs2894054 | 6p21.33 | Double case-control, 469cases, 390 women with persistent HR-HPV and 452 controls | Costa Rica (150) | - |
| MICA | 6p21.33 | GWAS, 1075 Cases and 4014 Controls | Sweden (96) | E5, E7 downregulate expression of MHC molecules (149) |
| SULF1, rs4737999 | 8q13.2-q13.3 | Double case-control, 469cases, 390 women with persistent HR-HPV and 452 control | Costa Rica (150) | - |
| IFNA1 | 9p21.3 | Cross-sectional, 148 patients | Thailand | E2, E6 downregulate IFNA1 (148) |
| IL2RA, rs2476491 | 10p15.1 | Double case-control, 141 cases, 38 HSIL and 176 controls | Portugal (151) | E6, E7 interferes with cytokines pathways (149, 152) |
| PRDX3, rs7082598 | 10q26.11 | Cross-sectional, 68 patients | China | E6, E7 downregulate PRDX3 (153) |
| C1RL, rs12227050 | 12p13.31 | Double case-control, 469cases, 390 women with persistent HR-HPV and 452 control | Costa Rica (154) | - |
| OAS3, rs12302655 | 12q24.13 | Double case-control, 469cases, 390 women with persistent HR-HPV and 452 control | Costa Rica (150) | - |
| DUT, rs3784621 | 15q21.1 | Double case-control, 469cases, 390 women with persistent HR-HPV and 452 control | Costa Rica (150) | - |
| TP53(p53), rs1042522 | 17p13.1 | Cross-sectional, 577 patients | USA (155) | E6 degrades p53 (156) |
| NLRP1, rs11651270 NLRP3, rs10754558 and IL18, rs1834481 | 17p13.2 | Case-control, 246 cases vs 310 controls, 12 SNPs in seven genes | Brazil (157) | - |
| TYMS, rs2342700 | 18p11.32 | Case-control, 65 cases vs 202 controls | Nigeria (158) | - |
| RPS19, rs2305809 | 19q13.2 | Case-control, 65 cases vs 202 controls | Nigeria (158) | - |
| IRF3, rs7251 | 19q13.33 | Double case-control, 469cases, 390 women with persistent HR-HPV and 452 control | Costa Rica (159) | E6 prevents IFN-α mRNA (160) |
| B) Genes involved in cervical disease progression | ||||
| EXO1, rs4149963 | 1q43 | Double case-control, 469cases, 390 women with persistent HR- HPV and 452 controls | Costa Rica (159) | - |
| TIPARP, rs2665390 | 3q25.3 | Case-control, 790 cases vs 717 controls | Algeria, Morocco, India, Thailand (161) | - |
| PIK3CA | 3q26.32 | Cross-sectional, 285 cases | Mexico, Guatemala, Venezuela (162) | E6, E7 increase APOBEC-mediated mutagenesis (163). |
| LAMP3 | 3q27.1 | Double case-control, 47 cases, 15 tissues with CIN and 5 tissue controls | Japan (164) | - |
| RFC4 | 3q27.3 | Case-control and Meta-analysis, 40 cases vs 20 controls | Brazil (165) | E1, E2 ORFs disruption interferes RFC4 (166) |
| POLN, rs17132382 | 4p16.3 | Double case-control, 416 cases, 356 women with persistent HR- HPV and 425 controls | Costa Rica (150) | E6 interacts with POLN (167) |
| MIR146A, rs2910164 | 5q33.3 | Case-control, 447 cases vs 443 controls | China (168) | E6 under-express miRNAs(169) |
| TNF | 6p21.33 | Descriptive, in vitro | USA (170) | E6 down-regulates TNF (171) |
| URG4 | 7p13 | Cross-sectional, 167 cc patients | China (172) | - |
| MYC | 8q24.2 | Descriptive, Case series, 1 cc patient | USA | E7 fusion causes MYC overexpression (173) |
| CDKN2A (p16) | 9p21.3 | Cross-sectional, 139 cases | Japan (174) | E7 inactivates Rb1 (p16 overexpression) (174) |
| TAP | 11q12.3 | Descriptive, in vitro | China (175) | E7 down-regulates TAP (171) |
| IFNG , rs11177074 | 12q15 | Double case-control, 416 cases, 356 women with persistent HR- HPV and 425 controls | Costa Rica (150) | E6 inhibit interferon related responses (171, 176). |
| MDM2 | 12q15 | Descriptive, in vitro | Italy | E2 interacts MDM2 ubiquitin ligase (177) |
| RB1 | 13q14.2 | Descriptive, in vitro | USA (178) | E7 binds and degrades pRb (178, 179). |
| CYP1A1 m2, rs1048943 | 15q24.1 | Case-control, 100 cases vs 100 controls | India (180) | - |
| TELO2, rs4786772 | 16p13.3 | Double case-control, 416 cases, 356 women with persistent HR- HPV and 425 controls | Costa Rica (154) | - |
| FANCA, rs2239359 | 16q24.3 | Double case-control, 469cases, 390 women with persistent HR- HPV and 452 controls | Costa Rica (159) | E6 promotes reprogramming of FANCA (181) |
| CYBA, rs7195830 | 16q24.3 | Double case-control, 469cases, 390 women with persistent HR- HPV and 452 controls | Costa Rica (159) | - |
| EVER1/EVER2, rs9893818 | 17q25.3 | Double case-control, 416 cases, 356 women with persistent HR- HPV and 425 controls | Costa Rica (150) | E7 binds zinc ions to prevent EVER1/2 and E5 binds to EVER1/2 (78) |
| FGFR-TK1 | 17q25.3 | Descriptive, Case series, 3 cc patients | USA and Brazil (182) | E6 induce expression of FGF-BP (182) |
It has been observed that, HR-HPV-DNA potentially integrates into more than 117 unique sites into the host genome to influence cervical carcinogenesis (109, 110). Several other genomic regions with changes in the number of DNA copies (copy number-altered regions or CNAs), common in solid tumors, have been confirmed by comparative genomic hybridization (CGH), Florescent 'In situ' Hybridization (FISH) and single nucleotide polymorphisms (SNPs), reflecting the important role of HPV infection and specific genomic alterations in cervical carcinogenesis (111).
Individuals with a Tp53 gene polymorphism in codon 72 (Arginine homozygosity) have been reported to be at a seven times higher risk for HPV associated cervical cancer development than the heterozygous genotype (37, 112). However, a pooled data analysis on 49 studies worldwide published in 2009, found no association between cervical cancer and the TP53 codon 72 polymorphism (113). The most frequently-mutated tumor suppressor gene in cervical cancer is believed to be Cystatin E/M, however, these results are not consistent in different study populations (114, 115).
Previous studies in cervical cancer molecular genetics using SNPs have shown that the accumulation of cellular genomic damage such as point mutations, gene amplifications, and LOH/MSI in both pre-cancerous and cancerous lesions occurs at rs13117307 at 4q12, rs8067378 at 17q12, rs4282438 and rs9277952 at 6p21.32 (89, 110). There were, however, no SNPs associated with cervical cancer risk in p21 rs1801270, BRIP1 rs2048718, and rs11079454 polymorphisms (116). Furthermore, the recurrent cellular genetic alterations in cervical cancer were observed only in primary mutational signatures, 1B and 2(APOBEC) (117) (118). Women who are carriers of genes or alleles that may affect the expression of immune molecules capable of HPV infection recognition are at increased risk for developing cervical cancer. However, variations in CD83, a marker of dendritic cell maturation that may assist the T cell response to HPV infection, have shown little or no influence on cervical cancer (119, 120). Contrarily, Yu et al (121) in 2009, confirmed an association between CD83 polymorphisms and cervical cancer susceptibility and suggested that polymorphisms in this gene and cervical disease association may depend on tumour histology.
Mutations in cyclin dependent kinase inhibitor, WAF1 have shown a positive association with cervical cancer susceptibility, although some studies, based on different study populations report contradictory findings (37, 122, 123). Somatic genomic mutations, notably copy number variations, in the genes PIK3CA, STK11, PTEN, TP53, and KRAS 4-7, have been associated with cervical cancer development (35, 63). Mutations, and/or polymorphisms in transporters associated with antigen processing genes, TAP-1 and TAP-2 were not associated with development of cervical cancer (124, 125). Although, Zoodzma et al (69) reported an increased risk of cervical cancer in individuals with allele 184 at the MICA locus (with a recessive effect), subsequent investigations have not been able to replicate this finding (126).
Certain heritable syndromes involving defects in the DNA damage repair system which present susceptibility to cervical cancer have been studied. For example, Fanconi anemia syndrome (genes include FANCA, FANCC, FANCL), is an inherited genetic disorder characterized by defects in DNA damage repair system. In addition, Mathew, (127) suggested an association between this syndrome and cervical cancer. However, in a subsequent study in a Swedish population, no association between Fanconi anemia and susceptibility to cervical cancer was shown (128). Enigmatically, however, in a study using FANCA gene deficient mice, Park et al (129) demonstrated susceptibility of HPV 16 E7-driven cervical cancer.
The ERAP1-575 gene on chromosome 5, and the TAP2-379, and TAP2-651 loci on chromosome 6, have been tested in Asian populations, and shown to be consistently associated with cervical cancer risk (130, 131). The rs799917 TT genotype in the BRCA1 gene has been associated with a significantly decreased risk of cervical cancer (89, 132). Some variants in the chemokine receptor-2A (CCR2A), a transcribed isoform of CD192, situated on chromosome 3p21, have demonstrated a protective effect against invasive cervical cancer development from squamous intraepithelial lesions (SIL) in Swedish, Portuguese, and South African Black and Mixed-Ancestry populations. It has also been described as a risk allele for high grade squamous intraepithelial lesions and cervical cancer development in healthy individuals. In other studies, however, the G46295A variant in CD192 was reported not to confer genetic susceptibility towards cervical cancer development (37, 133-135).
The activation of Caspase 8 (CASP8), represents an important initiating event in the death receptor-induced apoptosis gene. However, the deleted allele of the CASP8 polymorphism has been associated with decreased risk for cervical cancer in a Chinese population (136). Studies in African populations have not found any association between CASP8 polymorphisms and cervical cancer susceptibility (65).
Although, we have discussed numerous interactions of several genes, chromosomes, SNPs and the HPV oncoproteins E6 and E7, the exact mechanisms of interactions with the HIV co-infection, and the mechanisms by which the combined pro-oncogenic effects of HIV Tat proteins and HPV oncoproteins E6 and E7, further provoke additional genetic alterations in some women to influence the rate of cervical cancer progression is not yet known (54, 137).
iv) What is not known in host molecular genetics of cervical carcinoma?
There is lack of available research addressing some specific questions that need to be asked:
a) Why do variations at the MHC II locus increase the risk of cervical cancer? Although this has not been studied before in an HIV/HPV co-infected population, it does appear that the main host genetic susceptibility factors for cervical cancer may be related to the immune recognition of HPV-infected or HPV transformed cervical epithelial cells (138).
b) Do variations in the HLA II genes, *DRB1*DQB1 influence HIV positive cervical carcinogenesis (139)?
c) Are host molecular genetic polymorphisms at certain genomic loci more likely to influence cervical carcinogenesis in HIV/HPV co-infected women (83)? The immunogenetics of cervical carcinomas from HIV positive women has not yet been studied (140, 141).
d) What are the effects of LOH/MSI at 6p in HIV/HPV co-infected cervical carcinomas (96)?
e) What are the effects of HAART on the incidence and severity of cervical cancer in HIV/HPV co-infected women with regard to molecular genetic variations?
f) How do the anti-apoptotic effects of Protease Inhibitors (PI), influence cervical carcinogenesis (44)?
g) Can the significant genomic loci, candidate tumour suppressor genes and the biological pathways of the genetic framework of susceptibility or heritability to cervical cancer which have been indicated by the reported genome-wide association studies be elucidated (84, 126, 142)?
h) Can further research confirm whether susceptibility loci in one population are specifically replicated in another e.g. HLA, 4q12, 17q12 (80, 116, 143, 144)?
i) Why is the apparent effect of the P72R polymorphism in TP53 gene not consistent in different ethnicities (116)? This is despite the evidence that cervical cancer susceptibility loci in each ethnic group vary considerably.
j) Does HIV/HPV co-infection provoke additional host genetic alterations on chromosome 6p specifically at the HLA-II loci, DRBI and DQB1, to influence the rate of cervical disease development in HIV/HPV co-infected women (139)?
Overall, the evidence suggests that HPV persistent infection with oncogenic genotypes is a necessary, but not sufficient, risk factor for cervical carcinogenesis. We hypothesise that, HIV co-infection exacerbates and increases the rate of progression to invasive cervical cancer by promoting additional genetic alterations and mutations in chromosomal regions carrying tumour suppressor genes, apoptosis-related genes, DNA damage-repair genes, and cell cycle- regulatory genes by the cumulative oncogenic effects of the combined viruses. It is, however, not yet clear why only a small subset of HIV/HPV co-infected women progress rapidly to the invasive disease and others do not.
4. Conclusions and recommendations
To the best of our knowledge, this is the first study to analyse the HIV/HPV co-infection and correlation with host molecular genetics in the development of cervical cancer. Although there is limited published data on the interaction of the HIV Tat proteins and HPV oncoproteins, E6 and E7, and host molecular genetic susceptibility to cervical cancer progression, this review compiles the reports on the major host molecular genetic risk factors that have been shown to be associated with both rapidly progressing cervical cancer progression and susceptibility.
Our review has considered the gaps in knowledge for cervical cancer progression in general, and in HIV/HPV co-infected women, particularly. We have provided an updated literature review, which includes large number of genes and possible mutations for host molecular genetic susceptibility to cervical cancer development. We have discussed the influence of HIV/HPV co-infection, HIV Tat protein, HPV oncoproteins E6 and E7and host tumour suppressor genes in the disease time course and compared various published studies. Proper focus on high-risk populations for cervical cancer disease (e.g. HIV-infected women) should decrease the number of advanced cases of invasive cervical cancer by identification of early molecular genetic changes. This can be achieved by assessing molecular genetic risk factors using predictive testing (PT) for cervical disease development in HIV/HPV co-infected women (145, 146). In addition, the implementation of new genetic profiling and cervical cancer-screening programs for all women infected with HIV is envisaged (146).
At this stage we cannot point to one or a few genes that may affect susceptibility, severity or increase rate of progression of cervical cancer in the presence of HIV and HPV co-infection as many genes are involved in different molecular pathways. Generally, the genetic variations or mutations that affect host genes for the immune response against oncogenic HPV clearance, tumour suppressor genes, apoptosis-related genes, DNA damage-repair genes and cell cycle- regulatory genes are responsible for cervical cancer susceptibility and rapidly progressing disease. There is also inter-population or multi-population differences and a range of confounding host and viral factors, including environmental effects such as host behaviour and demographics. This may be why different genes have been found to be associated, or not associated, with cervical cancer development in different study populations.
In a South African population, we are currently investigating the hypothesis that HIV/HPV co-infection provokes additional host genetic alterations on chromosome 6p specifically at the HLA-II loci, DRBI and DQB1, to influence the rate of cervical disease development in HIV/HPV co-infected women (139). The impact of this work is expected to advance our current understanding of the interaction between HIV and HPV co-infections and rapidly invasive cervical cancer progression with regard to host molecular genetic and immunogenetic variations. Findings from this study will enlighten the primary goals of genetically-targeted therapy, in the prevention of disease, and the individualization of cervical cancer treatment (89). This will enable genetic profiling and individualization in prevention and treatment of invasive cervical cancer in women living with HIV/AIDS. It will also assist in the development of immunogenetic markers to detect cervical cancer development in early stages through severity of LOH/MSI. This work, therefore, recognises the urgent need to move away from the “one size fits all” generalization in prevention and treatment of cervical cancer (89).
Acknowledgements
This work is based on the research supported in part by the Postgraduate Academic Mobility for African Physician-Scientists (PAMAPS) organised by the University of Ibadan in Nigeria and funded by the European Union. We would like to thank Professor Tim Quinlan, from The Health Economics and HIV/AIDS Research Division (HEARD) at the University of KwaZulu-Natal, for his assistance in discussing and reviewing early drafts of the article.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ononogbu U, Almujtaba M, Modibbo F, Lawal I, Offiong R, Olaniyan O. et al. Cervical cancer risk factors among HIV-infected Nigerian women. BMC public health. 2013;13(1):582
2. Chambuso RS, Kaambo E, Stephan S. Observed Age Difference and Clinical Characteristics of Invasive Cervical Cancer Patients in Tanzania; A Comparison between HIV-Positive and HIV-Negative Women. Journal of Neoplasm. 2017:2 (3)
3. Chambuso RS, Shadrack S, Lidenge SJ, Mwakibete N, Medeiros RM. Influence of HIV/AIDS on Cervical Cancer: A Retrospective Study From Tanzania. Journal of Global Oncology. 2017;3(1):72-8
4. Adler DH, Wallace M, Bennie T, Abar B, Meiring TL, Williamson A-L. et al. Cumulative Impact of HIV and Multiple Concurrent Human Papillomavirus Infections on the Risk of Cervical Dysplasia. Advances in Virology. 2016;2016:5
5. Kriek JM, Jaumdally SZ, Masson L, Little F, Mbulawa Z, Gumbi PP. et al. Female genital tract inflammation, HIV co-infection and persistent mucosal Human Papillomavirus (HPV) infections. Virology. 2016;493:247-54
6. Moodley JR, Constant D, Hoffman M, Salimo A, Allan B, Rybicki E. et al. Human papillomavirus prevalence, viral load and pre-cancerous lesions of the cervix in women initiating highly active antiretroviral therapy in South Africa: a cross-sectional study. BMC cancer. 2009;9:275
7. Alves BM, Prellwitz IM, Siqueira JD, Meyrelles AR, Bergmann A, Seuanez HN. et al. The effect of human leukocyte antigen G alleles on human papillomavirus infection and persistence in a cohort of HIV-positive pregnant women from Brazil. Infect Genet Evol. 2015;34:339-43
8. Nyagol J, Leucci E, Onnis A, De Falco G, Tigli C, Sanseverino F. et al. The effects of HIV-1 Tat protein on cell cycle during cervical carcinogenesis. Cancer biology & therapy. 2006;5(6):684-90
9. Ghebre RG, Grover S, Xu MJ, Chuang LT, Simonds H. Cervical cancer control in HIV-infected women: Past, present and future. Gynecologic Oncology Reports. 2017;21:101-8
10. Kelly HA, Ngou J, Chikandiwa A, Sawadogo B, Gilham C, Omar T. et al. Associations of Human Papillomavirus (HPV) genotypes with high-grade cervical neoplasia (CIN2+) in a cohort of women living with HIV in Burkina Faso and South Africa. PloS one. 2017;12(3):e0174117
11. van Bogaert LJ. The impact of human immunodeficiency virus infection on cervical preinvasive and invasive neoplasia in South Africa. Ecancermedicalscience. 2013;7:334
12. Agorastos T, Chatzistamatiou K, Katsamagkas T, Koliopoulos G, Daponte A, Constantinidis T. et al. Primary Screening for Cervical Cancer Based on High-Risk Human Papillomavirus (HPV) Detection and HPV 16 and HPV 18 Genotyping, in Comparison to Cytology. PloS one. 2015;10(3):e0119755
13. Sundstrom K, Ploner A, Arnheim-Dahlstrom L, Eloranta S, Palmgren J, Adami HO. et al. Interactions Between High- and Low-Risk HPV Types Reduce the Risk of Squamous Cervical Cancer. Journal of the National Cancer Institute. 2015:107 (10)
14. Zhang X, Zhang L, Tian C, Yang L, Wang Z. Genetic variants and risk of cervical cancer: epidemiological evidence, meta-analysis and research review. BJOG: an international journal of obstetrics and gynaecology. 2014;121(6):664-74
15. Gokhale P, Kerkar S, Tongaonkar H, Chaudhari H, Warke H, Salvi V. et al. Variations in immunogenetics, human papillomavirus (HPV) infection & predisposition to cervical cancer in Indian women. Indian Journal of Medical Research. 2014;140:36-43
16. Williamson A-L. The Interaction between Human Immunodeficiency Virus and Human Papillomaviruses in Heterosexuals in Africa. Journal of Clinical Medicine. 2015;4(4):579
17. Passmore J-AS, Williamson A-L. Host Immune Responses Associated with Clearance or Persistence of Human Papillomavirus Infections. Current Obstetrics and Gynecology Reports. 2016;5(3):177-88
18. Pantanowitz L, Michelow P. Review of Human Immunodeficiency Virus (HIV) and squamous lesions of the uterine cervix. Diagnostic Cytopathology. 2011;39(1):65-72
19. Denny LA, Franceschi S, de Sanjosé S, Heard I, Moscicki AB, Palefsky J. Human Papillomavirus, Human Immunodeficiency Virus and Immunosuppression. Vaccine. 2012;30:F168-F74
20. Papasavvas E, Surrey LF, Glencross DK, Azzoni L, Joseph J, Omar T. et al. High-risk oncogenic HPV genotype infection associates with increased immune activation and T cell exhaustion in ART-suppressed HIV-1-infected women. OncoImmunology. 2016;5(5):e1128612
21. Rodriguez-Carunchio L, Soveral I, Steenbergen RD, Torne A, Martinez S, Fuste P. et al. HPV-negative carcinoma of the uterine cervix: a distinct type of cervical cancer with poor prognosis. BJOG: an international journal of obstetrics and gynaecology. 2015;122(1):119-27
22. Wang X, Wang G, Zhang L, Cong J, Hou J, Liu C. LncRNA PVT1 promotes the growth of HPV positive and negative cervical squamous cell carcinoma by inhibiting TGF-beta1. Cancer cell international. 2018;18:70
23. Wani K, Nair CK. Genetic alterations in cervical cancer. Indian journal of experimental biology. 2003;41(8):789-96
24. Kang S, Kim JW, Park NH, Song YS, Park SY, Kang SB. et al. Interleukin-1 Beta -511 Polymorphism and Risk of Cervical Cancer. Journal of Korean medical science. 2007;22(1):110-3
25. Lutkowska A, Roszak A, Lianeri M, Sowinska A, Sotiri E, Jagodzinski PP. Analysis of rs8067378 Polymorphism in the Risk of Uterine Cervical Cancer from a Polish Population and its Impact on Gasdermin B Expression. Mol Diagn Ther. 2017;21(2):199-207
26. Guzman VB, Yambartsev A, Goncalves-Primo A, Silva IDCG, Carvalho CRN, Ribalta JCL. et al. New approach reveals CD28 and IFNG gene interaction in the susceptibility to cervical cancer. Human molecular genetics. 2008;17(12):1838-44
27. Meys R, Purdie KJ, de Koning MN, Quint KD, Little AM, Baker F. et al. HLA Immunogenotype Determines Persistent Human Papillomavirus Virus Infection in HIV-Infected Patients Receiving Antiretroviral Treatment. The Journal of infectious diseases. 2016;213(11):1717-24
28. Ivansson EL, Juko-Pecirep I, Gyllensten UB. Interaction of immunological genes on chromosome 2q33 and IFNG in susceptibility to cervical cancer. Gynecologic oncology. 2010;116(3):544-8
29. Wei L-z, Wang H-l, Liu X, Lu Y-p, Xu F, Yuan J-q. et al. Meta-analysis on the relationship between HLA-DRBl gene polymorphism and cervical cancer in Chinese population. PloS one. 2014;9(2):e88439
30. Ou CY, Chang JG, Tseng HH, Wei HJ, Su TH, Hsu TY. et al. Analysis of microsatellite instability in cervical cancer. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 1999;9(1):67-71
31. Mazurenko N, Bliyev AY, Bidzhieva B, Peskov DY, Snigur N, Savinova E. et al. Loss of heterozygosity at chromosome 6 as a marker of early genetic alterations in cervical intraepithelial neoplasias and microinvasive carcinomas. Molecular Biology. 2006;40(3):385-95
32. Abdus-Salam A, Ogunnorin O, Abdus-Salam R. HIV Seroprevalence in Patients with Carcinoma of the Cervix in Ibadan, Nigeria. Ghana medical journal. 2008;42(4):141-3
33. Boccalon M, Tirelli U, Sopracordevole F, Vaccher E. Intra-epithelial and invasive cervical neoplasia during HIV infection. European journal of cancer. 1996;32(13):2212-7
34. Whitham HK, Hawes SE, Chu H, Oakes JM, Lifson AR, Kiviat NB. et al. A Comparison of the Natural History of HPV Infection and Cervical Abnormalities among HIV-Positive and HIV-Negative Women in Senegal, Africa. Cancer Epidemiology Biomarkers & Prevention. 2017
35. Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ. et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506(7488):371-5
36. Mehta AM, Mooij M, Brankovic I, Ouburg S, Morre SA, Jordanova ES. Cervical Carcinogenesis and Immune Response Gene Polymorphisms: A Review. J Immunol Res. 2017;2017:8913860
37. Chattopadhyay K. A comprehensive review on host genetic susceptibility to human papillomavirus infection and progression to cervical cancer. Indian Journal of Human Genetics. 2011;17(3):132-44
38. McBride AA, Warburton A. The role of integration in oncogenic progression of HPV-associated cancers. PLOS Pathogens. 2017;13(4):e1006211
39. Wani K, Nair CKK. Genetic alterations in cervical cancer. Indian Journal of Expcrimcntal Biology. 2003;41:789-96
40. PA L. The molecular genetics of cervical carcinoma. British journal of cancer. 1999;80(12):2008-18
41. de Freitas AC, Gurgel APAD, Chagas BS, Coimbra EC, do Amaral CMM. Susceptibility to cervical cancer: An overview. Gynecologic oncology. 2012;126(2):304-11
42. Al-Daraji WI, Smith JHF. Infection and Cervical Neoplasia: Facts and Fiction. International Journal of Clinical and Experimental Pathology. 2009;2(1):48-64
43. Arias-Pulido H, Joste N, Wheeler CM. Loss of heterozygosity on chromosome 6 in HPV-16 positive cervical carcinomas carrying the DRB1*1501-DQB1*0602 haplotype. Genes, chromosomes & cancer. 2004;40(4):277-84
44. Adefolaju GA, Theron KE, Hosie MJ. Effects of HIV protease, nucleoside/non-nucleoside reverse transcriptase inhibitors on Bax, Bcl-2 and apoptosis in two cervical cell lines. Biomedicine & Pharmacotherapy. 2014;68(2):241-51
45. Ramjee G, Moonsamy S, Abbai NS, Wand H. Individual and Population Level Impact of Key HIV Risk Factors on HIV Incidence Rates in Durban, South Africa. PloS one. 2016;11(4):e0153969
46. Louie KS, de Sanjose S, Mayaud P. Epidemiology and prevention of human papillomavirus and cervical cancer in sub-Saharan Africa: a comprehensive review. Tropical medicine & international health: TM & IH. 2009;14(10):1287-302
47. McDonald AC, Tergas AI, Kuhn L, Denny L, Wright TC Jr. Distribution of Human Papillomavirus Genotypes among HIV-Positive and HIV-Negative Women in Cape Town, South Africa. Frontiers in oncology. 2014;4:48
48. Investigators TST. Short-Course Antiretroviral Therapy in Primary HIV Infection. New England Journal of Medicine. 2013;368(3):207-17
49. Guidry JT, Scott RS. The interaction between human papillomavirus and other viruses. Virus research. 2017;231:139-47
50. de Jong A, van Poelgeest MIE, van der Hulst JM, Drijfhout JW, Fleuren GJ, Melief CJM. et al. Human Papillomavirus Type 16-Positive Cervical Cancer Is Associated with Impaired CD4+ T-Cell Immunity against Early Antigens E2 and E6. Cancer Research. 2004;64(15):5449-55
51. Moodley M, Mould S. Invasive cervical cancer and human immunodeficiency virus (HIV) infection in KwaZulu-Natal, South Africa. Journal of obstetrics and gynaecology: the journal of the Institute of Obstetrics and Gynaecology. 2005;25(7):706-10
52. Cubie HA. Diseases associated with human papillomavirus infection. Virology. 2013;445(1):21-34
53. Nicol AF, Pires AR, de Souza SR, Nuovo GJ, Grinsztejn B, Tristao A. et al. Cell-cycle and suppressor proteins expression in uterine cervix in HIV/HPV co-infection: comparative study by tissue micro-array (TMA). BMC cancer. 2008;8:289
54. Barillari G, Palladino C, Bacigalupo I, Leone P, Falchi M, Ensoli B. Entrance of the Tat protein of HIV-1 into human uterine cervical carcinoma cells causes upregulation of HPV-E6 expression and a decrease in p53 protein levels. Oncology Letters. 2016;12(4):2389-94
55. Vernon SD, Hart CE, Reeves WC, Icenogle JP. The HIV-1 tat protein enhances E2-dependent human papillomavirus 16 transcription. Virus research. 1993;27(2):133-45
56. Webster K, Parish J, Pandya M, Stern PL, Clarke AR, Gaston K. The human papillomavirus (HPV) 16 E2 protein induces apoptosis in the absence of other HPV proteins and via a p53-dependent pathway. Journal of Biological Chemistry. 2000;275(1):87-94
57. Hirbod T, Kimani J, Tjernlund A, Cheruiyot J, Petrova A, Ball TB. et al. Stable CD4 expression and local immune activation in the ectocervical mucosa of HIV-infected women. Journal of immunology. 2013;191(7):3948-54
58. Nkwanyana NN, Gumbi PP, Roberts L, Denny L, Hanekom W, Soares A. et al. Impact of human immunodeficiency virus 1 infection and inflammation on the composition and yield of cervical mononuclear cells in the female genital tract. Immunology. 2009;128(1 Suppl):e746-57
59. Bebell LM, Passmore JA, Williamson C, Mlisana K, Iriogbe I, van Loggerenberg F. et al. Relationship between levels of inflammatory cytokines in the genital tract and CD4+ cell counts in women with acute HIV-1 infection. The Journal of infectious diseases. 2008;198(5):710-4
60. Iuliano M, Mangino G, Chiantore MV, Zangrillo MS, Accardi R, Tommasino M. et al. Human Papillomavirus E6 and E7 oncoproteins affect the cell microenvironment by classical secretion and extracellular vesicles delivery of inflammatory mediators. Cytokine. 2017
61. Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436-44
62. Allavena P, Garlanda C, Borrello MG, Sica A, Mantovani A. Pathways connecting inflammation and cancer. Current opinion in genetics & development. 2008;18(1):3-10
63. Akram Husain R, Ramakrishnan V. A review of risk factors in the development of cervical malignancy. Cancer Sci Res Open Access. 2016;3(1):1-4
64. Nicol AF, Nuovo GJ, Salomao-Estevez A, Grinsztejn B, Tristao A, Russomano F. et al. Immune factors involved in the cervical immune response in the HIV/HPV co-infection. Journal of clinical pathology. 2008;61(1):84-8
65. Tan SC, Ankathil R. Genetic susceptibility to cervical cancer: role of common polymorphisms in apoptosis-related genes. Tumor Biology. 2015;36(9):6633-44
66. Williams VM, Filippova M, Soto U, Duerksen-Hughes PJ. HPV-DNA integration and carcinogenesis: putative roles for inflammation and oxidative stress. Future virology. 2011;6(1):45-57
67. Wong NS, Reidpath DD, Wong KH, Lee SS. A multilevel approach to assessing temporal change of CD4 recovery following HAART initiation in a cohort of Chinese HIV positive patients. Journal of Infection. 2015;70(6):676-8
68. Allen M, Kalantari M, Ylitalo N, Pettersson B, Hagmar B, Scheibenpflug L. et al. HLA DQ-DR haplotype and susceptibility to cervical carcinoma: indications of increased risk for development of cervical carcinoma in individuals infected with HPV 18. HLA. 1996;48(1):32-7
69. Zoodsma M, Nolte IM, Schipper M, Oosterom E, van der Steege G, de Vries EGE. et al. Analysis of the entire HLA region in susceptibility for cervical cancer: a comprehensive study. Journal of Medical Genetics. 2005;42(8):e49-e
70. Koko M, Suleiman SH, Abdallah MOE, Saad M, Ibrahim M. In-silico analysis of MHC genes in hereditary colorectal cancer shows identical by state SNP sharing affecting HLA-DQB1 binding groove. bioRxiv. 2016:040436
71. Zhang X, Lv Z, Yu H, Wang F, Zhu J. The HLA-DQB1 gene polymorphisms associated with cervical cancer risk: A meta-analysis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2015;73:58-64
72. Hardikar S, Johnson LG, Malkki M, Petersdorf EW, Galloway DA, Schwartz SM. et al. A population-based case-control study of genetic variation in cytokine genes associated with risk of cervical and vulvar cancers. Gynecologic oncology. 2015;139(1):90-6
73. Beskow AH, Gyllensten UB. Host genetic control of HPV 16 titer in carcinoma in situ of the cervix uteri. International journal of cancer Journal international du cancer. 2002;101(6):526-31
74. Stauss HJ, Davies H, Sadovnikova E, Chain B, Horowitz N, Sinclair C. Induction of cytotoxic T lymphocytes with peptides in vitro: identification of candidate T-cell epitopes in human papilloma virus. Proc Natl Acad Sci U S A. 1992;89(17):7871-5
75. Chen D, Hammer J, Lindquist D, Idahl A, Gyllensten U. A variant upstream of HLA-DRB1 and multiple variants in MICA influence susceptibility to cervical cancer in a Swedish population. Cancer medicine. 2014;3(1):190-8
76. Zhao X, Sun Q, Tian H, Cong B, Jiang X, Peng C. Loss of heterozygosity at 6p21 and HLA class I expression in esophageal squamous cell carcinomas in China. Asian Pacific journal of cancer prevention: APJCP. 2011;12(10):2741-5
77. Das Ghosh D, Mukhopadhyay I, Bhattacharya A, Roy Chowdhury R, Mandal NR, Roy S. et al. Impact of genetic variations and transcriptional alterations of HLA class I genes on cervical cancer pathogenesis. International Journal of Cancer. 2017;140(11):2498-508
78. Lazarczyk M, Cassonnet P, Pons C, Jacob Y, Favre M. The EVER proteins as a natural barrier against papillomaviruses: a new insight into the pathogenesis of human papillomavirus infections. Microbiology and Molecular Biology Reviews. 2009;73(2):348-70
79. Sudenga SL, Wiener HW, King CC, Rompalo AM, Cu-Uvin S, Klein RS. et al. Dense genotyping of immune-related loci identifies variants associated with clearance of HPV among HIV-positive women in the HIV epidemiology research study (HERS). PloS one. 2014;9(6):e99109
80. Chen D, Gyllensten U. Systematic investigation of contribution of genetic variation in the HLA-DP region to cervical cancer susceptibility. Carcinogenesis. 2014;35(8):1765-9
81. Zhang X, Lv Z, Yu H, Wang F, Zhu J. The HLA-DQB1 gene polymorphisms associated with cervical cancer risk: A meta-analysis. Biomedicine & Pharmacotherapy. 2015;73:58-64
82. Ivansson EL, Magnusson JJ, Magnusson PKE, Erlich HA, Gyllensten UB. MHC loci affecting cervical cancer risk: distinguishing the effects of HLA-DQB1 and non-HLA genes TNF, LTA, TAP1 and TAP2. Genes And Immunity. 2008;9:613
83. Engelmark M, Beskow A, Magnusson J, Erlich H, Gyllensten U. Affected sib-pair analysis of the contribution of HLA class I and class II loci to development of cervical cancer. Human molecular genetics. 2004;13(17):1951-8
84. Leo PJ, Madeleine MM, Wang S, Schwartz SM, Newell F, Pettersson-Kymmer U. et al. Defining the genetic susceptibility to cervical neoplasia—A genome-wide association study. PLOS Genetics. 2017;13(8):e1006866
85. Chen D, Gyllensten U. A cis-eQTL of HLA-DRB1 and a frameshift mutation of MICA contribute to the pattern of association of HLA alleles with cervical cancer. Cancer Medicine. 2014;3(2):445-52
86. Badano I, Stietz SM, Schurr TG, Picconi AM, Fekete D, Quintero IM. et al. Analysis of TNFα promoter SNPs and the risk of cervical cancer in urban populations of Posadas (Misiones, Argentina). Journal of Clinical Virology. 2012;53(1):54-9
87. Deshpande A, Nolan JP, White PS, Valdez YE, Hunt WC, Peyton CL. et al. TNF-α Promoter Polymorphisms and Susceptibility to Human Papillomavirus 16-Associated Cervical Cancer. The Journal of infectious diseases. 2005;191(6):969-76
88. Maciag PC, Villa LL. Genetic susceptibility to HPV infection and cervical cancer. Brazilian Journal of Medical and Biological Research. 1999;32:915-22
89. Chen X, Jiang J, Shen H, Hu Z. Genetic susceptibility of cervical cancer. Journal of Biomedical Research. 2011;25(3):155-64
90. Zheng H-T, Peng Z-H, Li S, He L. Loss of heterozygosity analyzed by single nucleotide polymorphism array in cancer. World Journal of Gastroenterology. 2005;11(43):6740-4
91. Harima Y, Harima K, Sawada S, Tanaka Y, Arita S, Ohnishi T. Loss of heterozygosity on chromosome 6p21.2 as a potential marker for recurrence after radiotherapy of human cervical cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2000;6(3):1079-85
92. Lo KC, Bailey D, Burkhardt T, Gardina P, Turpaz Y, Cowell JK. Comprehensive analysis of loss of heterozygosity events in glioblastoma using the 100K SNP mapping arrays and comparison with copy number abnormalities defined by BAC array comparative genomic hybridization. Genes, chromosomes & cancer. 2008;47(3):221-37
93. Zheng HT, Peng ZH, Li S, He L. Loss of heterozygosity analyzed by single nucleotide polymorphism array in cancer. World journal of gastroenterology: WJG. 2005;11(43):6740-4
94. Thiagalingam S, Foy RL, Cheng KH, Lee HJ, Thiagalingam A, Ponte JF. Loss of heterozygosity as a predictor to map tumor suppressor genes in cancer: molecular basis of its occurrence. Current opinion in oncology. 2002;14(1):65-72
95. Hunt JL. Chapter 5 - Loss of Heterozygosity A2 - Tubbs, Raymond R. In: (ed.) Stoler MH. Cell and Tissue Based Molecular Pathology. Philadelphia: Churchill Livingstone. 2009 p. 50-5
96. Chen D, Juko-Pecirep I, Hammer J, Ivansson E, Enroth S, Gustavsson I. et al. Genome-wide association study of susceptibility loci for cervical cancer. Journal of the National Cancer Institute. 2013;105(9):624-33
97. Ramos TDP, Amorim LMF. Molecular biology techniques for loss of heterozygosity detection: the glioma example. Jornal Brasileiro de Patologia e Medicina Laboratorial. 2015;51:189-96
98. Magnusson PK, Lichtenstein P, Gyllensten UB. Heritability of cervical tumours. International journal of cancer Journal international du cancer. 2000;88(5):698-701
99. Ivansson E. Contribution of Immunogenetic factors in susceptibility to cervical cancer [PhD Dissertation]. Uppsala University: Uppsala University. 2009
100. Borges ÁH, Dubrow R, Silverberg MJ. Factors contributing to risk for cancer among HIV-infected individuals, and evidence that earlier cART will alter this risk. Current opinion in HIV and AIDS. 2014;9(1):34-40
101. Genetic polymorphisms as predictive and prognostic biomarkers in gynecological cancers. a systematic review. 2012.
102. Shi Y, Li L, Hu Z, Li S, Wang S, Liu J. et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat Genet. 2013;45(8):918-22
103. Garner D. Clinical application of DNA ploidy to cervical cancer screening: A review. World Journal of Clinical Oncology. 2014;5(5):931-65
104. Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP. et al. The presence of extra chromosomes leads to genomic instability. Nature communications. 2016;7:10754
105. Danielsen HE, Pradhan M, Novelli M. Revisiting tumour aneuploidy — the place of ploidy assessment in the molecular era. Nature Reviews Clinical Oncology. 2015;13:291
106. Barh D, Gunduz M. Noninvasive Molecular Markers in Gynecologic Cancers: CRC Press; 2015.
107. Scotto L, Narayan G, Nandula SV, Subramaniyam S, Kaufmann AM, Wright JD. et al. Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Molecular Cancer. 2008;7:58 -
108. Jafari-Ghahfarokhi H, Moradi-Chaleshtori M, Liehr T, Hashemzadeh-Chaleshtori M, Teimori H, Ghasemi-Dehkordi P. Small supernumerary marker chromosomes and their correlation with specific syndromes. Advanced Biomedical Research. 2015;4:140
109. Liu Y, Lu Z, Xu R, Ke Y. Comprehensive mapping of the human papillomavirus (HPV) DNA integration sites in cervical carcinomas by HPV capture technology. Oncotarget. 2016;7(5):5852-64
110. Grozdanov P, Hadjidekova S, Dimova I, Nikolova I, Toncheva D, Ganchev G. et al. Characterization of genomic changes in the cervical pre-cancerous lesions and tumors induced by different types of human papillomaviruses. Virusdisease. 2016;27(3):271-6
111. Kuglik P, Smetana J, Vallova V, Moukova L, Kasikova K, Cvanova M. et al. Genome-wide screening of DNA copy number alterations in cervical carcinoma patients with CGH+SNP microarrays and HPV-FISH. International Journal of Clinical and Experimental Pathology. 2014;7(8):5071-82
112. Hu X, Zhang Z, Ma D, Huettner PC, Massad LS, Nguyen L. et al. TP53, MDM2, NQO1, and Susceptibility to Cervical Cancer. Cancer Epidemiology Biomarkers & Prevention. 2010;19(3):755-61
113. Klug SJ, Ressing M, Koenig J, Abba MC, Agorastos T, Brenna SMF. et al. TP53 codon 72 polymorphism and cervical cancer: a pooled analysis of individual data from 49 studies. The Lancet Oncology. 2009;10(8):772-84
114. Veena MS, Lee G, Keppler D, Mendonca MS, Redpath JL, Stanbridge EJ. et al. Inactivation of the cystatin E/M tumor suppressor gene in cervical cancer. Genes, chromosomes & cancer. 2008;47(9):740-54
115. Kloth JN, Oosting J, van Wezel T, Szuhai K, Knijnenburg J, Gorter A. et al. Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer. BMC Genomics. 2007;8:53
116. Martínez-Nava GA, Fernández-Niño JA, Madrid-Marina V, Torres-Poveda K. Cervical Cancer Genetic Susceptibility: A Systematic Review and Meta-Analyses of Recent Evidence. PloS one. 2016;11(7):e0157344
117. Litwin TR, Clarke MA, Dean M, Wentzensen N. Somatic Host Cell Alterations in HPV Carcinogenesis. Viruses. 2017:9 (8)
118. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV. et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415-21
119. Immuno-related polymorphisms and cervical cancer risk. The IARC multicentric case-control study. Public Library of Science. 2017 p. 1-13
120. Bodelon C, Madeleine MM, Johnson LG, Du Q, Malkki M, Petersdorf EW. et al. Genetic variation in CD83 and risks of cervical and vulvar cancers: a population-based case-control study. Gynecologic oncology. 2012;124(3):525-8
121. Yu KJ, Rader JS, Borecki I, Zhang Z, Hildesheim A. CD83 polymorphisms and cervical cancer risk. Gynecologic oncology. 2009;114(2):319-22
122. Kim YT, Zhao M. Aberrant Cell Cycle Regulation in Cervical Carcinoma. Yonsei Med J. 2005;46(5):597-613
123. Bahrami A, Hasanzadeh M, Shahidsales S, Farazestanian M, Hassanian SM, Moetamani Ahmadi M. et al. Genetic susceptibility in cervical cancer: From bench to bedside. Journal of Cellular Physiology. 2018;233(3):1929-39
124. Kordi Tamandani DM, Sobti RC, Shekari M, Husseini SA, Suri V. No association of TAP1 and TAP2 genes polymorphism with risk of cervical cancer in north Indian population. Journal of Assisted Reproduction and Genetics. 2009;26(4):173-8
125. Einstein MH, Leanza S, Chiu LG, Schlecht NF, Goldberg GL, Steinberg BM. et al. Genetic Variants in TAP Are Associated with High-Grade Cervical Neoplasia. Clinical Cancer Research. 2009;15(3):1019-23
126. Chen D, Cui T, Ek WE, Liu H, Wang H, Gyllensten U. Analysis of the genetic architecture of susceptibility to cervical cancer indicates that common SNPs explain a large proportion of the heritability. Carcinogenesis. 2015;36(9):992-8
127. Mathew CG. Fanconi anaemia genes and susceptibility to cancer. Oncogene. 2006;25:5875
128. Juko-Pecirep I, Ivansson EL, Gyllensten UB. Evaluation of Fanconi anaemia genes FANCA, FANCC and FANCL in cervical cancer susceptibility. Gynecologic oncology. 2011;122(2):377-81
129. Park JW, Shin MK, Lambert PF. High incidence of female reproductive tract cancers in FA-deficient HPV16-transgenic mice correlates with E7's induction of DNA damage response, an activity mediated by E7's inactivation of pocket proteins. Oncogene. 2014;33(26):3383-91
130. Mehta AM, Spaans VM, Mahendra NB, Osse EM, Vet JN, Purwoto G. et al. Differences in genetic variation in antigen-processing machinery components and association with cervical carcinoma risk in two Indonesian populations. Immunogenetics. 2015;67(5-6):267-75
131. Mehta AM, Osse M, Kolkman-Uljee S, Fleuren GJ, Jordanova ES. Molecular backgrounds of ERAP1 downregulation in cervical carcinoma. Analytical Cellular Pathology. 2015:2015
132. Zhou X, Han S, Wang S, Chen X, Dong J, Shi X. et al. Polymorphisms in HPV E6/E7 protein interacted genes and risk of cervical cancer in Chinese women: a case-control analysis. Gynecologic oncology. 2009;114(2):327-31
133. Chen Y, Liu X, Chen M, Shen J, Lu Y, Yu L. et al. CD192 gene variant and susceptibility to cervical cancer: a meta analysis. International Journal of Clinical and Experimental Medicine. 2015;8(6):9341-7
134. Chatterjee K, Dandara C, Hoffman M, Williamson A-L. CCR2-V64I polymorphism is associated with increased risk of cervical cancer but not with HPV infection or pre-cancerous lesions in African women. BMC cancer. 2010;10:278 -
135. Coelho A, Matos A, Catarino R, Pinto D, Sousa H, Pereira D. et al. The Influence of Chemokine Receptor CCR2 Genotypes in the Route to Cervical Carcinogenesis. Gynecologic and Obstetric Investigation. 2007;64(4):208-12
136. Xiao M-S, Chang L, Li W-L, Du Y-S, Pan Y, Zhang D-F. et al. Genetic Polymorphisms of the CASP8 Gene Promoter May Not Be Associated with Colorectal Cancer in Han Chinese from Southwest China. PloS one. 2013;8(7):e67577
137. Chen D, Enroth S, Ivansson E, Gyllensten U. Pathway analysis of cervical cancer genome-wide association study highlights the MHC region and pathways involved in response to infection. Human molecular genetics. 2014;23(22):6047-60
138. Engelmark M, Beskow A, Magnusson J, Erlich H, Gyllensten U. Affected sib-pair analysis of the contribution of HLA class I and class II loci to development of cervical cancer. Human molecular genetics. 2004;13(17):1951-8
139. Chen D, Enroth S, Liu H, Sun Y, Wang H, Yu M. et al. Pooled analysis of genome-wide association studies of cervical intraepithelial neoplasia 3 (CIN3) identifies a new susceptibility locus. Oncotarget. 2016;7(27):42216-24
140. Isaakidis P, Pimple S, Varghese B, Khan S, Mansoor H, Ladomirska J. et al. HPV infection, cervical abnormalities, and cancer in HIV-infected women in Mumbai, India: 12-month follow-up. International journal of women's health. 2013;5:487-94
141. Madeleine MM, Brumback B, Cushing-Haugen KL, Schwartz SM, Daling JR, Smith AG. et al. Human Leukocyte Antigen Class II and Cervical Cancer Risk: A Population-Based Study. The Journal of infectious diseases. 2002;186(11):1565-74
142. Hu X, Zhang Z, Ma D, Huettner PC, Massad LS, Nguyen L. et al. TP53, MDM2, NQO1, and susceptibility to cervical cancer. Cancer Epidemiology and Prevention Biomarkers. 2010;19(3):755-61
143. Shi Y, Li L, Hu Z, Li S, Wang S, Liu J. et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nature genetics. 2013;45(8):918-22
144. Qiu H, Tang W, Yin P, Cheng F, Wang L. Cytotoxic T-lymphocyte-associated antigen-4 polymorphisms and susceptibility to cervical cancer: A meta-analysis. Molecular medicine reports. 2013;8(6):1785-94
145. Bruwer Z, Algar U, Vorster A, Fieggen K, Davidson A, Goldberg P. et al. Predictive genetic testing in children: constitutional mismatch repair deficiency cancer predisposing syndrome. Journal of genetic counseling. 2014;23(2):147-55
146. Bruwer Z, Futter M, Ramesar R. Communicating cancer risk within an African context: experiences, disclosure patterns and uptake rates following genetic testing for Lynch syndrome. Patient education and counseling. 2013;92(1):53-60
147. Su T-H, Chang T-Y, Lee Y-J, Chen C-K, Liu H-F, Chu C-C. et al. CTLA-4 gene and susceptibility to human papillomavirus-16-associated cervical squamous cell carcinoma in Taiwanese women. Carcinogenesis. 2007;28(6):1237-40
148. Sunthamala N, Thierry F, Teissier S, Pientong C, Kongyingyoes B, Tangsiriwatthana T. et al. E2 Proteins of High Risk Human Papillomaviruses Down-Modulate STING and IFN-κ Transcription in Keratinocytes. PloS one. 2014;9(3):e91473
149. Westrich JA, Warren CJ, Pyeon D. Evasion of host immune defenses by human papillomavirus. Virus research. 2017;231:21-33
150. Wang SS, Gonzalez P, Yu K, Porras C, Li Q, Safaeian M. et al. Common genetic variants and risk for HPV persistence and progression to cervical cancer. PloS one. 2010;5(1):e8667
151. Craveiro R, Bravo I, Catarino R, Teixeira AL, Sousa H, Pereira D. et al. The role of p73 G4C14-to-A4T14 polymorphism in the susceptibility to cervical cancer. DNA and cell biology. 2012;31(2):224-9
152. Scott ME, Shvetsov YB, Thompson PJ, Hernandez BY, Zhu X, Wilkens LR. et al. Cervical Cytokines and Clearance of Incident Human Papillomavirus Infection: Hawaii HPV Cohort Study. International journal of cancer Journal international du cancer. 2013;133(5):1187-96
153. Hu J-X, Gao QUN, Li L. Peroxiredoxin 3 is a novel marker for cell proliferation in cervical cancer. Biomedical Reports. 2013;1(2):228-30
154. Safaeian M, Hildesheim A, Gonzalez P, Yu K, Porras C, Li Q. et al. Single nucleotide polymorphisms in the PRDX3 and RPS19 and risk of HPV persistence and cervical precancer/cancer. PloS one. 2012;7(4):e33619
155. Hu X, Zhang Z, Ma D, Huettner P, Massad LS, Nguyen L. et al. TP53, MDM2, NQO1 and susceptibility to Cervical Cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19(3):755-61
156. Kardani K, Bolhassani A. HPV Proteins and Their Functions. HPV Infections: Diagnosis, Prevention, and Treatment. 2018:8
157. Pontillo A, Bricher P, Leal VNC, Lima S, Souza PRE, Crovella S. Role of inflammasome genetics in susceptibility to HPV infection and cervical cancer development. Journal of medical virology. 2016;88(9):1646-51
158. Famooto A, Almujtaba M, Dareng E, Akarolo-Anthony S, Ogbonna C, Offiong R. et al. RPS19 and TYMS SNPs and Prevalent High Risk Human Papilloma Virus Infection in Nigerian Women. PloS one. 2013;8(6):e66930
159. Wang SS, Bratti MC, Rodriguez AC, Herrero R, Burk RD, Porras C. et al. Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. The Journal of infectious diseases. 2009;199(1):20-30
160. Bordignon V, Di Domenico EG, Trento E, D'Agosto G, Cavallo I, Pontone M. et al. How Human Papillomavirus Replication and Immune Evasion Strategies Take Advantage of the Host DNA Damage Repair Machinery. Viruses. 2017;9(12):390
161. McKay J, Tenet V, Franceschi S, Chabrier A, Gheit T, Gaborieau V. et al. Immuno-related polymorphisms and cervical cancer risk: The IARC multicentric case-control study. PloS one. 2017;12(5):e0177775
162. Lou H, Villagran G, Boland JF, Im KM, Polo S, Zhou W. et al. Genome Analysis of Latin American Cervical Cancer: Frequent Activation of the PIK3CA Pathway. Clinical Cancer Research. 2015
163. Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell reports. 2014;7(6):1833-41
164. Kanao H, Enomoto T, Kimura T, Fujita M, Nakashima R, Ueda Y. et al. Overexpression of <em>LAMP3/TSC403/DC-LAMP</em> Promotes Metastasis in Uterine Cervical Cancer. Cancer Research. 2005;65(19):8640-5
165. Mine KL, Shulzhenko N, Yambartsev A, Rochman M, Sanson GF, Lando M. et al. Gene network reconstruction reveals cell cycle and antiviral genes as major drivers of cervical cancer. Nature communications. 2013;4:1806
166. Tsakogiannis D, Gortsilas P, Kyriakopoulou Z, Ruether IGA, Dimitriou TG, Orfanoudakis G. et al. Sites of disruption within E1 and E2 genes of HPV16 and association with cervical dysplasia. Journal of medical virology. 2015;87(11):1973-80
167. Ganguly N, Parihar SP. Human papillomavirus E6 and E7 oncoproteins as risk factors for tumorigenesis. Journal of biosciences. 2009;34(1):113-23
168. Yue C, Wang M, Ding B, Wang W, Fu S, Zhou D. et al. Polymorphism of the pre-miR-146a is associated with risk of cervical cancer in a Chinese population. Gynecologic oncology. 2011;122(1):33-7
169. Gómez-Gómez Y, Organista-Nava J, Gariglio P. Deregulation of the miRNAs Expression in Cervical Cancer: Human Papillomavirus Implications. BioMed Research International. 2013;2013:407052
170. Filippova M, Song H, Connolly JL, Dermody TS, Duerksen-Hughes PJ. The human papillomavirus 16 E6 protein binds to tumor necrosis factor (TNF) R1 and protects cells from TNF-induced apoptosis. J Biol Chem. 2002;277(24):21730-9
171. Habbous S, Pang V, Xu W, Amir E, Liu G. Human papillomavirus and host genetic polymorphisms in carcinogenesis: A systematic review and meta-analysis. Journal of Clinical Virology. 2014;61(2):220-9
172. Zhang L, Huang H, Zhang L, Hou T, Wu S, Huang Q. et al. URG4 overexpression is correlated with cervical cancer progression and poor prognosis in patients with early-stage cervical cancer. BMC cancer. 2014;14:885
173. Yuan H, Krawczyk E, Blancato J, Albanese C, Zhou D, Wang N. et al. HPV positive neuroendocrine cervical cancer cells are dependent on Myc but not E6/E7 viral oncogenes. Scientific Reports. 2017;7:45617
174. Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T. Expression Status of p16 Protein Is Associated with Human Papillomavirus Oncogenic Potential in Cervical and Genital Lesions. The American Journal of Pathology. 1998;153(6):1741-8
175. Li W, Deng XM, Wang CX, Zhang X, Zheng GX, Zhang J. et al. Down-regulation of HLA class I antigen in human papillomavirus type 16 E7 expressing HaCaT cells: correlate with TAP-1 expression. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2010;20(2):227-32
176. Mine KL, Shulzhenko N, Yambartsev A, Rochman M, Sanson GFO, Lando M. et al. Gene network reconstruction reveals cell cycle and antiviral genes as major drivers of cervical cancer. Nature communications. 2013;4:1806 -
177. Gammoh N, Gardiol D, Massimi P, Banks L. The Mdm2 Ubiquitin Ligase Enhances Transcriptional Activity of Human Papillomavirus E2. Journal of virology. 2009;83(3):1538-43
178. White EA, Münger K, Howley PM. High-risk human papillomavirus E7 proteins target PTPN14 for degradation. MBio. 2016;7(5):e01530-16
179. Buitrago-Pérez Á, Garaulet G, Vázquez-Carballo A, Paramio JM, García-Escudero R. Molecular Signature of HPV-Induced Carcinogenesis: pRb, p53 and Gene Expression Profiling. Current Genomics. 2009;10(1):26-34
180. Jain V, Ratre YK, Amle D, Mishra PK, Patra PK. Polymorphism of CYP1A1 gene variants rs4646903 and rs1048943 relation to the incidence of cervical cancer in Chhattisgarh. Environmental Toxicology and Pharmacology. 2017;52:188-92
181. Chlon TM, Hoskins EE, Mayhew CN, Wikenheiser-Brokamp KA, Davies SM, Mehta P. et al. High-risk human papillomavirus E6 protein promotes reprogramming of Fanconi anemia patient cells through repression of p53 but does not allow for sustained growth of induced pluripotent stem cells. Journal of virology. 2014;88(19):11315-26
182. Carneiro BA, Elvin JA, Kamath SD, Ali SM, Paintal AS, Restrepo A. et al. FGFR3-TACC3: A novel gene fusion in cervical cancer. Gynecologic Oncology Reports. 2015;13:53-6
Author contact
![]() Corresponding author: rchambusocom
Corresponding author: rchambusocom